6EEE
 
 | | X-ray crystal structure of Pf-M17 in complex with inhibitor (6k) and regulatory zinc ion | | Descriptor: | (1R,2r,3S,5R,7R)-N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)ethyl]tricyclo[3.3.1.1~3,7~]decane-2-carboxamide, 1,2-ETHANEDIOL, CARBONATE ION, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2018-08-13 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Hydroxamic Acid Inhibitors Provide Cross-Species Inhibition of Plasmodium M1 and M17 Aminopeptidases.
J. Med. Chem., 62, 2019
|
|
6KNI
 
 | |
6EH7
 
 | |
6JTO
 
 | | Crystal structure of HLA-C05 in complex with a tumor mut10m peptide | | Descriptor: | 10-mer peptide, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Bai, P, Zhou, Q, Wei, P, Lei, Y. | | Deposit date: | 2019-04-11 | | Release date: | 2020-04-15 | | Last modified: | 2021-05-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational discovery of a cancer neoepitope harboring the KRAS G12D driver mutation.
Sci China Life Sci, 2021
|
|
6EJE
 
 | |
6EIX
 
 | | Crystal structure of the kinase domain of the Q207E mutant of ACVR1 (ALK2) in complex with a 2-aminopyridine inhibitor K02288 | | Descriptor: | 1,2-ETHANEDIOL, 3-[6-amino-5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenol, Activin receptor type-1 | | Authors: | Williams, E.P, Canning, P, Sanvitale, C.E, Krojer, T, Allerston, C.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2017-09-19 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the kinase domain of the Q207E mutant of ACVR1 (ALK2) in complex with K02288
To be published
|
|
6EJ8
 
 | |
6JWM
 
 | |
6ESD
 
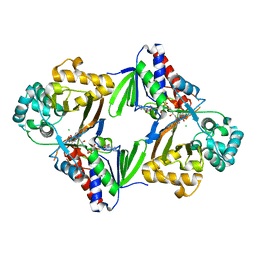 | | Crystal structure of L-tryptophan oxidase VioA from Chromobacterium violaceum | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent L-tryptophan oxidase VioA | | Authors: | Lai, H.E, Morgan, M, Moore, S, Freemont, P. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A semi-synthetic strategy for derivatization of the violacein natural product scaffold
Biorxiv, 2017
|
|
6E58
 
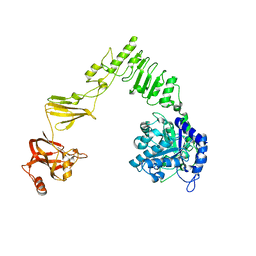 | | Crystal structure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) | | Descriptor: | CALCIUM ION, Secreted Endo-beta-N-acetylglucosaminidase (EndoS) | | Authors: | Klontz, E.H, Trastoy, B, Gunther, S, Guerin, M.E, Sundberg, E.J. | | Deposit date: | 2018-07-19 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular Basis of Broad SpectrumN-Glycan Specificity and Processing of Therapeutic IgG Monoclonal Antibodies by Endoglycosidase S2.
ACS Cent Sci, 5, 2019
|
|
6JZS
 
 | | Structure of the Manganese Protoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan in complex with Pyridine | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Stanfield, J.K, Omura, K, Matsumoto, A, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-05-03 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6K0V
 
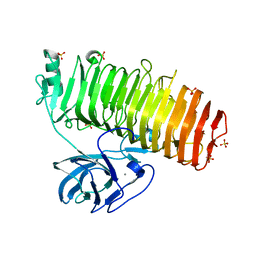 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with tetrasaccharides | | Descriptor: | Alpha-1,3-glucanase, CALCIUM ION, SULFATE ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6E6V
 
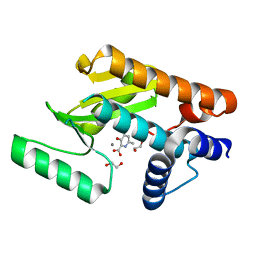 | | The N-terminal domain of PA endonuclease from the influenza H1N1 virus in complex with 3-hydroxy-6-methyl-4-oxo-1,4-dihydropyridine-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxy-6-methyl-4-oxo-1,4-dihydropyridine-2-carboxylic acid, MANGANESE (II) ION, ... | | Authors: | Dick, B.L, Morrison, C.N, Credille, C.V, Cohen, S.M. | | Deposit date: | 2018-07-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SAR Exploration of Tight-Binding Inhibitors of Influenza Virus PA Endonuclease.
J.Med.Chem., 62, 2019
|
|
6E8R
 
 | |
6EA1
 
 | | X-ray crystal structure of Pf-M1 in complex with inhibitor (6da) and catalytic zinc ion | | Descriptor: | (2R)-2,3,3,3-tetrafluoro-N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)ethyl]propanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Drinkwater, N, McGowan, S. | | Deposit date: | 2018-08-02 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.815 Å) | | Cite: | Hydroxamic Acid Inhibitors Provide Cross-Species Inhibition of Plasmodium M1 and M17 Aminopeptidases.
J. Med. Chem., 62, 2019
|
|
6E8S
 
 | | Structure of the iMango-III aptamer bound to TO1-Biotin | | Descriptor: | 4-[(3-{2-[(2-methoxyethyl)amino]-2-oxoethyl}-1,3-benzothiazol-3-ium-2-yl)methyl]-1-methylquinolin-1-ium, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Trachman, R.J, Ferre-D'Amare, A.R. | | Deposit date: | 2018-07-31 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure and functional reselection of the Mango-III fluorogenic RNA aptamer.
Nat. Chem. Biol., 15, 2019
|
|
6E9W
 
 | | Crystal structure of Rock1 with a pyridinylbenzamide based inhibitor | | Descriptor: | N-[(2,3-dihydro-1,4-benzodioxin-5-yl)methyl]-4-(pyridin-4-yl)benzamide, Rho-associated protein kinase 1, SULFATE ION | | Authors: | Judge, R.A, Hobson, A.D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Identification of Selective Dual ROCK1 and ROCK2 Inhibitors Using Structure Based Drug Design.
J. Med. Chem., 61, 2018
|
|
6K6M
 
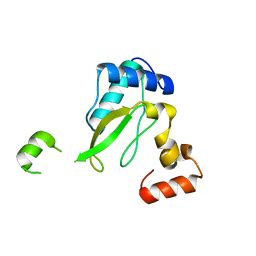 | | Crystal structure of HIV-2 Nef protein | | Descriptor: | Protein Nef | | Authors: | Hirao, K, Andrews, S, Kuroki, K, Kusaka, H, Tadokoro, T, Kita, S, Ose, T, Rowland-Jones, S, Maenaka, K. | | Deposit date: | 2019-06-04 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.072 Å) | | Cite: | Structure of HIV-2 Nef Reveals Features Distinct from HIV-1 Involved in Immune Regulation.
Iscience, 23, 2020
|
|
6EA9
 
 | |
6EAL
 
 | |
6EAZ
 
 | |
6EB9
 
 | |
6K8K
 
 | | Crystal structure of Arabidopsis thaliana BIC2-CRY2 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wang, X, Ma, L, Guan, Z, Yin, P. | | Deposit date: | 2019-06-12 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6EAI
 
 | |
6ECF
 
 | | Vlm2 thioesterase domain with genetically encoded 2,3-diaminopropionic acid bound with a dodecadepsipeptide, space group P1 | | Descriptor: | Vlm2, dodecadepsipeptide | | Authors: | Alonzo, D.A, Huguenin-Dezot, N, Heberlig, G.W, Mahesh, M, Nguyen, D.P, Dornan, M.H, Boddy, C.N, Chin, J.W, Schmeing, T.M. | | Deposit date: | 2018-08-07 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Trapping biosynthetic acyl-enzyme intermediates with encoded 2,3-diaminopropionic acid.
Nature, 565, 2019
|
|
