6BLF
 
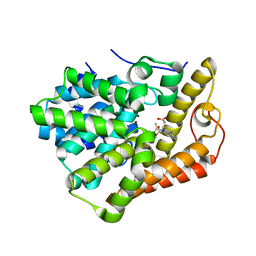 | | PDE2 complexed with 2-[6-fluoro-8-methylsulfonyl-9-[(1R)-1-[4-(trifluoromethyl)phenyl]ethyl]-1,2,3,4-tetrahydrocarbazol-1-yl]acetic acid | | Descriptor: | MAGNESIUM ION, ZINC ION, [(1S)-6-fluoro-8-(methylsulfonyl)-9-{(1R)-1-[4-(trifluoromethyl)phenyl]ethyl}-2,3,4,9-tetrahydro-1H-carbazol-1-yl]acetic acid, ... | | Authors: | Su, H.P. | | Deposit date: | 2017-11-10 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Hydrolase complex
To Be Published
|
|
5AGJ
 
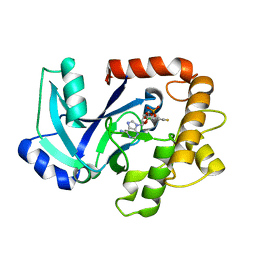 | | Crystal structure of the LeuRS editing domain of Candida albicans in complex with the adduct AN2690-AMP | | Descriptor: | POTENTIAL CYTOSOLIC LEUCYL TRNA SYNTHETASE, [(6-AMINO-9H-PURIN-9-YL)-[5-FLUORO-1,3-DIHYDRO-1-HYDROXY-2,1-BENZOXABOROLE]-4'YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Zhao, H, Palencia, A, Seiradake, E, Ghaemi, Z, Luthey-Schulten, Z, Cusack, S, Martinis, S.A. | | Deposit date: | 2015-02-02 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of the Resistance Mechanism of a Benzoxaborole Inhibitor Reveals Insight Into the Leucyl-tRNA Synthetase Editing Mechanism.
Acs Chem.Biol., 10, 2015
|
|
7T7M
 
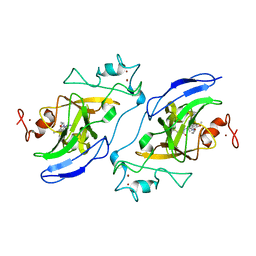 | | Structure of human GLP SET-domain (EHMT1) in complex with covalent inhibitor (Compound 1) | | Descriptor: | Histone-lysine N-methyltransferase EHMT1, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)prop-2-enamide, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)propanamide, ... | | Authors: | Park, K.-S, Kumar, P. | | Deposit date: | 2021-12-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of the First-in-Class G9a/GLP Covalent Inhibitors.
J.Med.Chem., 65, 2022
|
|
5UZC
 
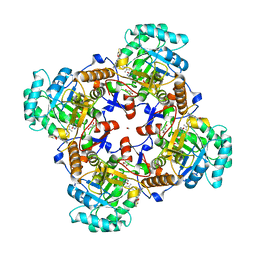 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P221 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-26 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P221
To Be Published
|
|
5V06
 
 | | Crystal structure of human exonuclease 1 Exo1 (WT) in complex with 5' recessed-end DNA (rIV) | | Descriptor: | DNA (5'-D(*CP*GP*CP*TP*AP*GP*TP*CP*GP*AP*CP*AP*T)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*TP*AP*GP*CP*G)-3'), Exonuclease 1, ... | | Authors: | Shi, Y, Beese, L.S. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Interplay of catalysis, fidelity, threading, and processivity in the exo- and endonucleolytic reactions of human exonuclease I.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6BQ0
 
 | | Structure of human monoacylglycerol lipase bound to a covalent inhibitor | | Descriptor: | 1-({(1R,5S,6r)-6-[1-(4-fluorophenyl)-1H-pyrazol-3-yl]-3-azabicyclo[3.1.0]hexane-3-carbonyl}oxy)pyrrolidine-2,5-dione, Monoglyceride lipase | | Authors: | Pandit, J. | | Deposit date: | 2017-11-27 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Trifluoromethyl Glycol Carbamates as Potent and Selective Covalent Monoacylglycerol Lipase (MAGL) Inhibitors for Treatment of Neuroinflammation.
J. Med. Chem., 61, 2018
|
|
7ET7
 
 | | Co-crystal structure of Human Nicotinamide N-methyltransferase (NNMT) with tricyclic small molecule inhibitor JBSNF-000028 | | Descriptor: | 10-methyl-1,10-diazatricyclo[6.3.1.0^{4,12}]dodeca-4,6,8(12)-trien-11-imine, Nicotinamide N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Swaminathan, S, Gosu, R, Birudukota, S, Kandan, S, Vaithilingam, K. | | Deposit date: | 2021-05-12 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Novel tricyclic small molecule inhibitors of Nicotinamide N-methyltransferase for the treatment of metabolic disorders.
Sci Rep, 12, 2022
|
|
7SER
 
 | | PRMT5/MEP50 with compound 30 bound | | Descriptor: | (2M)-2-{4-[4-(aminomethyl)-1-oxo-1,2-dihydrophthalazin-6-yl]-1-methyl-1H-pyrazol-5-yl}-1-benzothiophene-3-carbonitrile, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, ... | | Authors: | Gunn, R.J, Thomas, N.C, Lawson, J.D, Ivetac, A, Smith, C.R, Kulyk, S, Marx, M.A. | | Deposit date: | 2021-10-01 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Fragment-Based Discovery of MRTX1719, a Synthetic Lethal Inhibitor of the PRMT5•MTA Complex for the Treatment of MTAP -Deleted Cancers.
J.Med.Chem., 65, 2022
|
|
5AE9
 
 | | Crystal structure of mouse PI3 kinase delta in complex with GSK2292767 | | Descriptor: | N-[5-[4-(5-{[(2R,6S)-2,6-DIMETHYL-4-MORPHOLINYL]METHYL}-1,3-OXAZOL-2-YL)-1H-INDAZOL-6-YL]-2-(METHYLOXY)-3-PYRIDINYL]METHANESULFONAMIDE, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Down, K.D, Amour, A, Baldwin, I.R, Cooper, A.W.J, Deakin, A.M, Felton, L.M, Guntrip, S.B, Hardy, C, Harrison, Z.A, Jones, K.L, Jones, P, Keeling, S.E, Le, J, Livia, S, Lucas, F, Lunniss, C.J, Parr, N.J, Robinson, E, Rowland, P, Smith, S, Thomas, D.A, Vitulli, G, Washio, Y, Hamblin, N. | | Deposit date: | 2015-08-26 | | Release date: | 2015-09-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Optimization of Novel Indazoles as Highly Potent and Selective Inhibitors of Phosphoinositide 3-Kinase Gamma for the Treatment of Respiratory Disease.
J.Med.Chem., 58, 2015
|
|
7FG9
 
 | | Alpha-1,2-glucosyltransferase_UDP_tll1591 | | Descriptor: | Glycosyl transferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Su, J.Y. | | Deposit date: | 2021-07-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis for glucosylsucrose synthesis by a member of the alpha-1,2-glucosyltransferase family
Acta Biochim.Biophys.Sin., 54, 2022
|
|
6FDI
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-226 | | Descriptor: | 1,2-ETHANEDIOL, 1-[2-[4-[(4~{a}~{S},8~{a}~{R})-4-(3,4-dimethoxyphenyl)-1-oxidanylidene-4~{a},5,8,8~{a}-tetrahydrophthalazin-2-yl]piperi din-1-yl]-2-oxidanylidene-ethyl]-4,4-dimethyl-piperidine-2,6-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2017-12-24 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-226
To be published
|
|
5YAL
 
 | | Ferulic acid esterase from Streptomyces cinnamoneus at 1.5 A resolution | | Descriptor: | ACETATE ION, Esterase, GLYCEROL, ... | | Authors: | Tamura, H, Uraji, M, Mizohata, E, Ogawa, K, Inoue, T, Hatanaka, T. | | Deposit date: | 2017-09-01 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Loop of Streptomyces Feruloyl Esterase Plays an Important Role in the Enzyme's Catalyzing the Release of Ferulic Acid from Biomass.
Appl. Environ. Microbiol., 84, 2018
|
|
5UPP
 
 | | Crystal structure of human fumarate hydratase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fumarate hydratase, mitochondrial | | Authors: | Rangel, V.L, Ajalla, M.A, Rustiguel, J.K, Pereira de Padua, R.A, Nonato, M.C. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural, biochemical and biophysical characterization of recombinant human fumarate hydratase.
Febs J., 2019
|
|
8XBB
 
 | | The substrate binding protein of an ABC transporter in complex with beta-1,3-xylobiose | | Descriptor: | XylA, beta-D-xylopyranose-(1-3)-beta-D-xylopyranose | | Authors: | Zhao, F, Sun, H.N, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2023-12-06 | | Release date: | 2024-12-11 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Complete xylan utilization pathway and regulation mechanisms involved in marine algae degradation by cosmopolitan marine and human gut microbiota.
Isme J, 19, 2025
|
|
6C0D
 
 | |
6EVC
 
 | | Structure of E282Q A. niger Fdc1 in complex with pentafluoro-cinnamic acid | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, David, L, Payne, K.A.P. | | Deposit date: | 2017-11-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | The role of conserved residues in Fdc decarboxylase in prenylated flavin mononucleotide oxidative maturation, cofactor isomerization, and catalysis.
J. Biol. Chem., 293, 2018
|
|
5HRP
 
 | | HIV Integrase Catalytic Domain containing F185K + A124T mutations complexed with GSK0002 | | Descriptor: | (2S)-tert-butoxy[1-(3,4-difluorobenzyl)-6-methyl-4-(5-methyl-3,4-dihydro-2H-chromen-6-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]acetic acid, 1,2-ETHANEDIOL, CACODYLATE ION, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2016-01-24 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
5VCX
 
 | | CRYSTAL STRUCTURE OF HUMAN MYT1 KINASE DOMAIN (UNTREATED) IN COMPLEX WITH SARACATINIB | | Descriptor: | 1,2-ETHANEDIOL, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, N-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-7-[2-(4-METHYLPIPERAZIN-1-YL)ETHOXY]-5-(TETRAHYDRO-2H-PYRAN-4-YLOXY)QUINAZOLIN-4-AMINE | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
6EZG
 
 | | Torpedo californica AChE in complex with indolic multi-target directed ligand | | Descriptor: | 1-(7-chloranyl-4-methoxy-1~{H}-indol-5-yl)-3-[1-(phenylmethyl)piperidin-4-yl]propan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Santoni, G, Lalut, J, Karila, D, Lecoutey, C, Davis, A, Nachon, F, Sillman, I, Sussman, J, Weik, M, Maurice, T, Dallemagne, P, Rochais, C. | | Deposit date: | 2017-11-15 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel multitarget-directed ligands targeting acetylcholinesterase and sigma1receptors as lead compounds for treatment of Alzheimer's disease: Synthesis, evaluation, and structural characterization of their complexes with acetylcholinesterase.
Eur J Med Chem, 162, 2018
|
|
4YC4
 
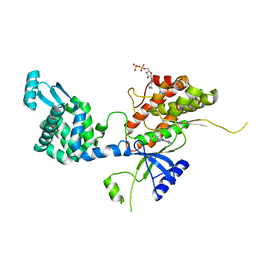 | | Crystal structure of phosphatidyl inositol 4-kinase II alpha in complex with nucleotide analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha,Lysozyme,Phosphatidylinositol 4-kinase type 2-alpha, [(1S,3S,4S)-3-(6-amino-9H-purin-9-yl)bicyclo[2.2.1]hept-1-yl]methanol | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2015-02-19 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The high-resolution crystal structure of phosphatidylinositol 4-kinase II beta and the crystal structure of phosphatidylinositol 4-kinase II alpha containing a nucleoside analogue provide a structural basis for isoform-specific inhibitor design.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5J4D
 
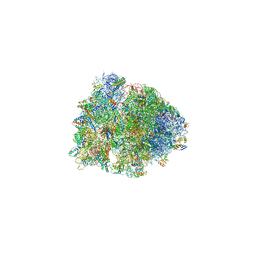 | |
6I82
 
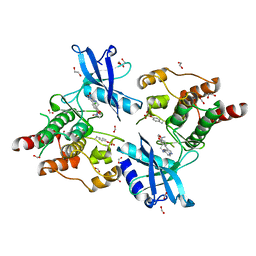 | | Crystal structure of partially phosphorylated RET V804M tyrosine kinase domain complexed with PDD00018412 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Burschowsky, D, Seewooruthun, C, Bayliss, R, Carr, M.D, Echalier, A, Jordan, A.M. | | Deposit date: | 2018-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Optimization of wt-RET/KDR-Selective Inhibitors of RETV804MKinase.
Acs Med.Chem.Lett., 11, 2020
|
|
5OA6
 
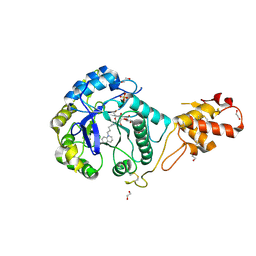 | | Crystal structure of ScGas2 in complex with compound 12 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-3,5-bis(oxidanyl)-6-[4-(3-quinolin-1-ium-1-ylpropyl)-1,2,3-triazol-1-yl]oxan-4-yl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-oxane-3,4,5-triol, 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2, ... | | Authors: | Delso, I, Valero-Gonzalez, J, Gomollon-Bel, F, Castro-Lopez, J, Fang, W, Navratilova, I, Van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-21 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
7C66
 
 | | Crystal structure of beta-glycosides-binding protein of ABC transporter in a closed state bound to cellobiose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
5NY6
 
 | | Carbonic Anhydrase II Inhibitor RA12 | | Descriptor: | 4-chloranyl-~{N}-[(1~{R})-1-(2-hydroxyphenyl)ethyl]-3-sulfamoyl-benzamide, 4-chloranyl-~{N}-[(1~{S})-1-(2-hydroxyphenyl)ethyl]-3-sulfamoyl-benzamide, Carbonic anhydrase 2, ... | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
