6STC
 
 | |
7AH1
 
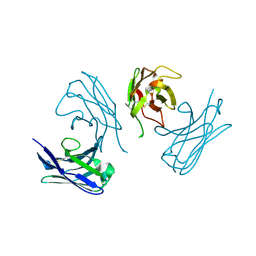 | | L19 diabody fragment from immunocytokine L19-IL2 | | Descriptor: | Anti-(ED-B) scFV | | Authors: | Ongaro, T, Guarino, S.R, Scietti, L, Palamini, M, Wulhfard, S, Villa, A, Neri, D, Forneris, F. | | Deposit date: | 2020-09-23 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inference of molecular structure for characterization and improvement of clinical grade immunocytokines.
J.Struct.Biol., 213, 2021
|
|
6STH
 
 | |
7AVC
 
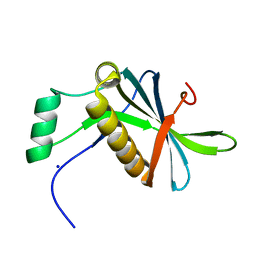 | | DoBi scaffold based on PIH1D1 N-terminal domain | | Descriptor: | GLYCEROL, PIH1 domain-containing protein 1, SODIUM ION | | Authors: | Kolenko, P, Pham, N.P, Pavlicek, J, Mikulecky, P, Schneider, B. | | Deposit date: | 2020-11-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Protein Binder (ProBi) as a New Class of Structurally Robust Non-Antibody Protein Scaffold for Directed Evolution.
Viruses, 13, 2021
|
|
6SRU
 
 | | Structure of Ig-like V-type domian of mouse Programmed cell death 1 ligand 1 (PD-L1) | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Magiera-Mularz, K, Sala, D, Grudnik, P, Holak, T.A. | | Deposit date: | 2019-09-06 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Human and mouse PD-L1: similar molecular structure, but different druggability profiles.
Iscience, 24, 2021
|
|
5MJ5
 
 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetichonokiol derivative 3 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[4-oxidanyl-3-[3-(phenylmethyl)phenyl]phenyl]prop-2-enoic acid, LYS-HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP-SER, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-11-30 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
5MFS
 
 | | The crystal structure of E. coli Aminopeptidase N in complex with 7-amino-4-phenyl-5,7,8,9-tetrahydrobenzocyclohepten-6-one | | Descriptor: | Aminopeptidase N, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Peng, G, Olieric, V, McEwen, A.G, Schmitt, C, Albrecht, S, Cavarelli, J, Tarnus, C. | | Deposit date: | 2016-11-18 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Insight into the remarkable affinity and selectivity of the aminobenzosuberone scaffold for the M1 aminopeptidases family based on structure analysis.
Proteins, 85, 2017
|
|
5MGD
 
 | | STRUCTURE OF E298Q-BETA-GALACTOSIDASE FROM ASPERGILLUS NIGER IN COMPLEX WITH 6-Galactosyl-lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Rico-Diaz, A, Ramirez-Escudero, M, Vizoso Vazquez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2016-11-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural features of Aspergillus niger beta-galactosidase define its activity against glycoside linkages.
FEBS J., 284, 2017
|
|
7AM3
 
 | | Crystal structure of Peptiligase mutant - M222P | | Descriptor: | GLYCEROL, SULFATE ION, Subtilisin BPN' | | Authors: | Rozeboom, H.J, Janssen, D.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | From thiol-subtilisin to omniligase: Design and structure of a broadly applicable peptide ligase.
Comput Struct Biotechnol J, 19, 2021
|
|
6SWG
 
 | |
5MGM
 
 | |
7B0T
 
 | | Crystal structure of MLLT1 YEATS domain T3 mutant in complex with benzimidazole-amide based compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 3-iodanyl-4-methyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Ni, X, Chaikuad, A, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-21 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and Inhibitor Binding Characterization of Oncogenic MLLT1 Mutants.
Acs Chem.Biol., 16, 2021
|
|
5MGS
 
 | | Human receptor NKR-P1 in deglycosylated form, extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor subfamily B member 1 | | Authors: | Skalova, T, Blaha, J, Stransky, J, Koval, T, Hasek, J, Yuguang, Z, Harlos, K, Vanek, O, Dohnalek, J. | | Deposit date: | 2016-11-22 | | Release date: | 2018-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Nat Commun, 13, 2022
|
|
6SWX
 
 | | Leishmania major methionyl-tRNA synthetase in complex with an allosteric inhibitor | | Descriptor: | METHIONINE, Putative methionyl-tRNA synthetase, methyl 2-[[6-[[3,4-bis(fluoranyl)phenyl]amino]-1-methyl-pyrazolo[3,4-d]pyrimidin-4-yl]amino]ethanoate | | Authors: | Robinson, D.A, Torrie, L.S, Shepherd, S.M, De Rycker, M, Thomas, M.G, Wyatt, P.G. | | Deposit date: | 2019-09-24 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of an Allosteric Binding Site in Kinetoplastid Methionyl-tRNA Synthetase.
Acs Infect Dis., 6, 2020
|
|
6ST3
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with 4-hydroxy-N-(4-phenoxybenzyl)-2-(1H-pyrazol-1-yl)pyrimidine-5-carboxamide | | Descriptor: | 4-oxidanyl-~{N}-[(4-phenoxyphenyl)methyl]-2-pyrazol-1-yl-pyrimidine-5-carboxamide, Egl nine homolog 1, FORMIC ACID, ... | | Authors: | Chowdhury, R, Holt-Martyn, J.P, Schofield, C.J. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.426 Å) | | Cite: | Structure-Activity Relationship and Crystallographic Studies on 4-Hydroxypyrimidine HIF Prolyl Hydroxylase Domain Inhibitors.
Chemmedchem, 15, 2020
|
|
7B10
 
 | | Crystal structure of MLLT1 YEATS domain T1 mutant in complex with benzimidazole-amide based compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 3-iodanyl-4-methyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, IODIDE ION, ... | | Authors: | Chaikuad, A, Ni, X, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-23 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure and Inhibitor Binding Characterization of Oncogenic MLLT1 Mutants.
Acs Chem.Biol., 16, 2021
|
|
5MHF
 
 | | Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin. | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mannosyl-oligosaccharide glucosidase, ... | | Authors: | Hill, J.C, Caputo, A.T, Roversi, P, Zitzmann, N. | | Deposit date: | 2016-11-24 | | Release date: | 2017-12-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting Endoplasmic Reticulum alpha-Glucosidase I with a Single-Dose Iminosugar Treatment Protects against Lethal Influenza and Dengue Virus Infections.
J.Med.Chem., 2020
|
|
7AM6
 
 | |
6STZ
 
 | |
7AQF
 
 | |
7AM8
 
 | | Crystal structure of Omniligase mutant W189F | | Descriptor: | ACRYLIC ACID, CHLORIDE ION, HISTIDINE, ... | | Authors: | Rozeboom, H.J, Janssen, D.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | From thiol-subtilisin to omniligase: Design and structure of a broadly applicable peptide ligase.
Comput Struct Biotechnol J, 19, 2021
|
|
5MMA
 
 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ379 (compound 5'g) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Maskell, D.P, Pye, V.E, Cherepanov, P. | | Deposit date: | 2016-12-09 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-Guided Optimization of HIV Integrase Strand Transfer Inhibitors.
J. Med. Chem., 60, 2017
|
|
7AM4
 
 | |
5MMR
 
 | | Crystal Structure of CK2alpha with N-((2-chloro-[1,1'-biphenyl]-4-yl)methyl)butane-1,4-diamine bound | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, PHOSPHATE ION, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K, Iegre, J, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2016-12-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A fragment-based approach leading to the discovery of a novel binding site and the selective CK2 inhibitor CAM4066.
Bioorg. Med. Chem., 25, 2017
|
|
6SYN
 
 | | Crystal structure of Y. pestis penicillin-binding protein 3 | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2S)-2-carboxy-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, ACETATE ION, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Pankov, G, Hunter, W.N, Dawson, A. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | The structure of penicillin-binding protein 3 from Yersinia pestis
To Be Published
|
|
