6KVN
 
 | |
5WIK
 
 | | JAK2 Pseudokinase in complex with BI-D1870 | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-5,7-dimethyl-8-(3-methylbutyl)-7,8-dihydropteridin-6(5H)-one, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5WIM
 
 | | JAK2 Pseudokinase in complex with AT9283 | | Descriptor: | 1-cyclopropyl-3-{3-[5-(morpholin-4-ylmethyl)-1H-benzimidazol-2-yl]-1H-pyrazol-4-yl}urea, Tyrosine-protein kinase JAK2 | | Authors: | Li, Q, Eck, M.J, Li, K, Park, E. | | Deposit date: | 2017-07-19 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Structural Characterization of ATP-Site Ligands for the Wild-Type and V617F Mutant JAK2 Pseudokinase Domain.
ACS Chem. Biol., 14, 2019
|
|
5WJO
 
 | | Crystal structure of the unliganded PG90 TCR | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PG90 TCR alpha chain, ... | | Authors: | Shahine, A, Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A molecular basis of human T cell receptor autoreactivity toward self-phospholipids.
Sci Immunol, 2, 2017
|
|
5I5R
 
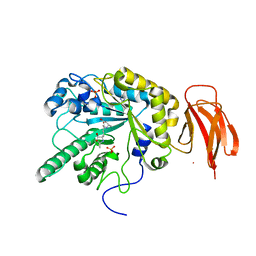 | | Crystal structure of a bacterial fucosidase with iminocyclitol (2S,3S,4R,5S)-3,4-dihydroxy-2-ethynyl-5-methylpyrrolidine | | Descriptor: | (2S,3S,4R,5S,2'S,3'S,4'R,5'S)-2,2'-(butane-1,4-diyl)bis(5-methylpyrrolidine-3,4-diol), Alpha-L-fucosidase, IMIDAZOLE, ... | | Authors: | Wright, D.W, Davies, G.J. | | Deposit date: | 2016-02-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Will be disclosed later
To Be Published
|
|
5I7I
 
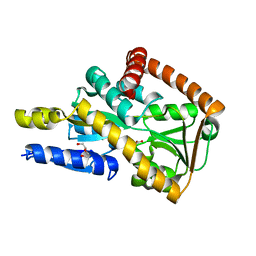 | | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157) in complex with co-crystallized 3-hydroxybenzoate | | Descriptor: | 3-HYDROXYBENZOIC ACID, DI(HYDROXYETHYL)ETHER, TRAP solute Binding Protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Hogle, S.L, Dupont, C.L, Almo, S.C. | | Deposit date: | 2016-02-17 | | Release date: | 2017-01-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of a marine metagenome TRAP solute binding protein specific for aromatic acid ligands (Sorcerer II Global Ocean Sampling Expedition, unidentified microbe, locus tag GOS_1523157) in complex with co-purified 4-hydroxybenzoate
To Be Published
|
|
5I8L
 
 | | Crystal structure of the full-length cell wall-binding module of Cpl7 mutant R223A | | Descriptor: | GLYCEROL, Lysozyme | | Authors: | Bernardo-Garcia, N, Silva-Martin, N, Uson, I, Hermoso, J.A. | | Deposit date: | 2016-02-19 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Deciphering how Cpl-7 cell wall-binding repeats recognize the bacterial peptidoglycan.
Sci Rep, 7, 2017
|
|
6L9B
 
 | | X-ray structure of synthetic GB1 domain with mutations K10(DVA), T11A | | Descriptor: | Immunoglobulin G-binding protein G | | Authors: | Penmatsa, A, Chatterjee, J, Khatri, B, Majumder, P. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Increasing protein stability by engineering the n -> pi * interaction at the beta-turn.
Chem Sci, 11, 2020
|
|
5I9Q
 
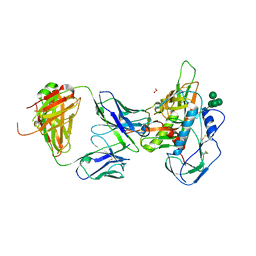 | | Crystal structure of 3BNC55 Fab in complex with 426c.TM4deltaV1-3 gp120 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC55 Fab heavy chain, ... | | Authors: | Scharf, L, Chen, C, Bjorkman, P.J. | | Deposit date: | 2016-02-20 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for germline antibody recognition of HIV-1 immunogens.
Elife, 5, 2016
|
|
5WL0
 
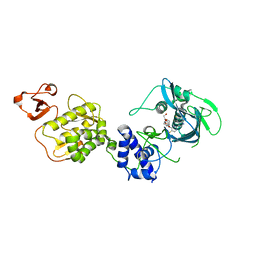 | | Co-crystal structure of Influenza A H3N2 PB2 (241-741) bound to VX-787 | | Descriptor: | (2S,3S)-3-[[5-fluoranyl-2-(5-fluoranyl-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-4-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, Polymerase basic protein 2 | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2017-07-25 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for therapeutic inhibition of influenza A polymerase PB2 subunit.
Sci Rep, 7, 2017
|
|
6L1J
 
 | | Crystal structure of barley exohydrolaseI W434A mutant in complex with 4'-nitrophenyl thiolaminaritrioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4'-NITROPHENYL-S-(BETA-D-GLUCOPYRANOSYL)-(1-3)-(3-THIO-BETA-D-GLUCOPYRANOSYL)-(1-3)-BETA-D-GLUCOPYRANOSIDE, ACETATE ION, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-09-29 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6KOI
 
 | | Crystal structure of SNX11-PXe domain in dimer form. | | Descriptor: | Sorting nexin-11 | | Authors: | Xu, T, Xu, J, Liu, J. | | Deposit date: | 2019-08-11 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular Basis for PI(3,5)P2Recognition by SNX11, a Protein Involved in Lysosomal Degradation and Endosome Homeostasis Regulation.
J.Mol.Biol., 432, 2020
|
|
6KOZ
 
 | | Crystal structure of two domain M1 zinc metallopeptidase E323 mutant bound to L-Leucine amino acid | | Descriptor: | LEUCINE, SODIUM ION, ZINC ION, ... | | Authors: | Agrawal, R, Kumar, A, Kumar, A, Makde, R.D. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the unusual substrate specificity of unique two-domain M1 metallopeptidase.
Int.J.Biol.Macromol., 147, 2020
|
|
5X1K
 
 | | Vanillate/3-O-methylgallate O-demethylase, LigM, 3-O-methylgallate complex form | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methoxy-4,5-bis(oxidanyl)benzoic acid, ... | | Authors: | Harada, A, Senda, T. | | Deposit date: | 2017-01-26 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a new O-demethylase from Sphingobium sp. strain SYK-6
FEBS J., 284, 2017
|
|
5WLI
 
 | | Crystal Structure of H-2Db with the GAP501 peptide (SQL) | | Descriptor: | Beta-2-microglobulin, GAP50 peptide, H-2 class I histocompatibility antigen, ... | | Authors: | Gras, S, Farenc, C, Josephs, T, Rossjohn, J. | | Deposit date: | 2017-07-27 | | Release date: | 2017-11-15 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A T Cell Receptor Locus Harbors a Malaria-Specific Immune Response Gene.
Immunity, 47, 2017
|
|
5WNB
 
 | | Structure of antibody 3D3 bound to the linear epitope of RSV G | | Descriptor: | ACETIC ACID, Major surface glycoprotein G, ZINC ION, ... | | Authors: | Fedechkin, S.O, George, N.L, Wolff, J.T, Kauvar, L.M, DuBois, R.M. | | Deposit date: | 2017-07-31 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of respiratory syncytial virus G antigen bound to broadly neutralizing antibodies.
Sci Immunol, 3, 2018
|
|
6KRH
 
 | | Structural basis for domain rotation during adenylation of active site K123 and fragment library screening against NAD+ -dependent DNA ligase from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase A, ... | | Authors: | Ramachandran, R, Shukla, A, Afsar, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Salt bridges at the subdomain interfaces of the adenylation domain and active-site residues of Mycobacterium tuberculosis NAD + -dependent DNA ligase A (MtbLigA) are important for the initial steps of nick-sealing activity.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5WNW
 
 | | Chaperone Spy bound to Im7 6-45 ensemble | | Descriptor: | CHLORIDE ION, Colicin-E7 immunity protein, IMIDAZOLE, ... | | Authors: | Horowitz, S, Salmon, L, Koldewey, P, Ahlstrom, L.S, Martin, R, Xu, Q, Afonine, P.V, Trievel, R.C, Brooks, C.L, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
6KRQ
 
 | |
6LCD
 
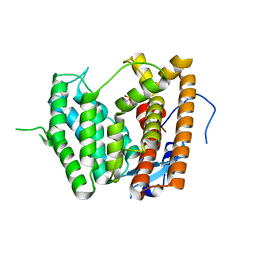 | |
6KSC
 
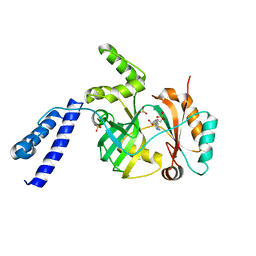 | |
6LCW
 
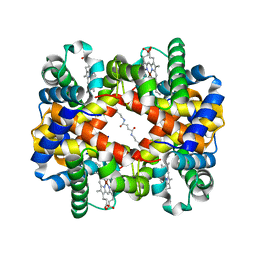 | | Crosslinked alpha(Ni)-beta(Ni) human hemoglobin A in the T quaternary structure at 95 K: Dark | | Descriptor: | BUT-2-ENEDIAL, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Shibayama, N, Park, S.Y, Ohki, M, Sato-Tomita, A. | | Deposit date: | 2019-11-20 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Direct observation of ligand migration within human hemoglobin at work.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5IFM
 
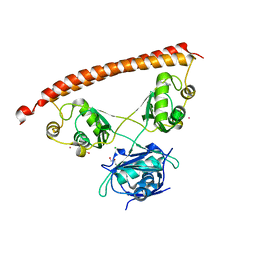 | | Human NONO (p54nrb) Homodimer | | Descriptor: | CHLORIDE ION, GLYCEROL, Non-POU domain-containing octamer-binding protein, ... | | Authors: | Knott, G.J, Bond, C.S. | | Deposit date: | 2016-02-26 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A crystallographic study of human NONO (p54(nrb)): overcoming pathological problems with purification, data collection and noncrystallographic symmetry.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6KQC
 
 | |
6KRK
 
 | |
