6JT5
 
 | | Crystal structure of PQQ doamin of Pyranose Dehydrogenase from Coprinopsis cinerea: apo-from | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular PQQ-dependent sugar dehydrogenase, ... | | Authors: | Takeda, K, Ishida, T, Yoshida, M, Samejima, M, Ohno, H, Igarashi, K, Nakamura, N. | | Deposit date: | 2019-04-09 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the Catalytic and CytochromebDomains in a Eukaryotic Pyrroloquinoline Quinone-Dependent Dehydrogenase.
Appl.Environ.Microbiol., 85, 2019
|
|
1FXH
 
 | | MUTANT OF PENICILLIN ACYLASE IMPAIRED IN CATALYSIS WITH PHENYLACETIC ACID IN THE ACTIVE SITE | | Descriptor: | 2-PHENYLACETIC ACID, CALCIUM ION, PENICILLIN ACYLASE | | Authors: | Alkema, W.B, Hensgens, C.M, Kroezinga, E.H, de Vries, E, Floris, R, van der Laan, J.M, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2000-09-26 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Characterization of the beta-lactam binding site of penicillin acylase of Escherichia coli by structural and site-directed mutagenesis studies.
Protein Eng., 13, 2000
|
|
2AVT
 
 | | Crystal structure of the beta subunit from DNA polymerase of Streptococcus pyogenes | | Descriptor: | DNA polymerase III beta subunit | | Authors: | Argiriadi, M.A, Goedken, E.R, Bruck, I, O'donnell, M, Kuriyan, J. | | Deposit date: | 2005-08-30 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a DNA polymerase sliding clamp from a Gram-positive bacterium.
Bmc Struct.Biol., 6, 2006
|
|
3DU8
 
 | | Crystal structure of GSK-3 beta in complex with NMS-869553A | | Descriptor: | (7S)-2-(2-aminopyrimidin-4-yl)-7-(2-fluoroethyl)-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one, Glycogen synthase kinase-3 beta | | Authors: | Bossi, R.T. | | Deposit date: | 2008-07-17 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | First Cdc7 kinase inhibitors: pyrrolopyridinones as potent and orally active antitumor agents. 2. Lead discovery.
J.Med.Chem., 52, 2009
|
|
6GJO
 
 | |
1FZ5
 
 | |
6GKF
 
 | |
1FYZ
 
 | | METHANE MONOOXYGENASE HYDROXYLASE, FORM II REDUCED BY SOAKING | | Descriptor: | CALCIUM ION, FE (II) ION, METHANE MONOOXYGENASE COMPONENT A, ... | | Authors: | Whittington, D.A, Lippard, S.J. | | Deposit date: | 2000-10-03 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of the soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath) demonstrating geometrical variability at the dinuclear iron active site.
J.Am.Chem.Soc., 123, 2001
|
|
1G06
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT V149S | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-05 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
4DHS
 
 | | Small-molecule inhibitors of 14-3-3 protein-protein interactions from virtual screening | | Descriptor: | (2-{2-[(3,5-dichlorophenyl)amino]-2-oxoethoxy}phenyl)phosphonic acid, 14-3-3 PROTEIN SIGMA, CHLORIDE ION, ... | | Authors: | Thiel, P, Roeglin, L, Kohlbacher, O, Ottmann, C. | | Deposit date: | 2012-01-30 | | Release date: | 2013-07-31 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions.
Chem.Commun.(Camb.), 49, 2013
|
|
1FZH
 
 | | METHANE MONOOXYGENASE HYDROXYLASE, FORM II PRESSURIZED WITH XENON GAS | | Descriptor: | CALCIUM ION, FE (III) ION, METHANE MONOOXYGENASE COMPONENT A, ... | | Authors: | Whittington, D.A, Rosenzweig, A.C, Frederick, C.A, Lippard, S.J. | | Deposit date: | 2000-10-03 | | Release date: | 2001-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase.
Biochemistry, 40, 2001
|
|
4LS3
 
 | | THE crystal STRUCTURE OF HELICOBACTER PYLORI CEUE(HP1561)/NI-HIS COMPL | | Descriptor: | HISTIDINE, NICKEL (II) ION, Nickel (III) ABC transporter, ... | | Authors: | Salamina, M, Shaik, M.M, Cendron, L, Zanotti, G. | | Deposit date: | 2013-07-22 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Helicobacter pylori periplasmic receptor CeuE (HP1561) modulates its nickel affinity via organic metallophores.
Mol.Microbiol., 91, 2014
|
|
1G0S
 
 | |
2B1L
 
 | |
4HGM
 
 | | Shark IgNAR Variable Domain | | Descriptor: | 1,2-ETHANEDIOL, ACETYL GROUP, Serum albumin, ... | | Authors: | Olland, A, Kovalenko, O.V, King, D, Svenson, K. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Atypical Antigen Recognition Mode of a Shark Immunoglobulin New Antigen Receptor (IgNAR) Variable Domain Characterized by Humanization and Structural Analysis.
J.Biol.Chem., 288, 2013
|
|
1G0L
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT T152V | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2000-10-06 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
5ZI8
 
 | | Crystal structure of the PadR-family transcriptional regulator Rv3488 of Mycobacterium tuberculosis H37Rv in complex with cadmium ion | | Descriptor: | CADMIUM ION, Transcriptional regulator | | Authors: | Meera, K, pal, R.K, Arora, A, Biswal, B.K. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional characterization of the transcriptional regulator Rv3488 ofMycobacterium tuberculosisH37Rv.
Biochem. J., 475, 2018
|
|
4HEC
 
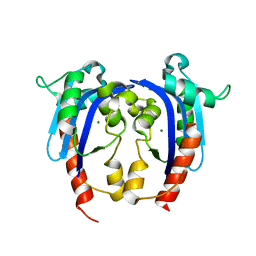 | |
1B47
 
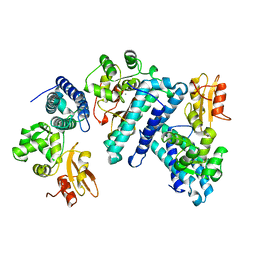 | | STRUCTURE OF THE N-TERMINAL DOMAIN OF CBL IN COMPLEX WITH ITS BINDING SITE IN ZAP-70 | | Descriptor: | CALCIUM ION, CBL | | Authors: | Meng, W, Sawasdikosol, S, Burakoff, S.J, Eck, M.J. | | Deposit date: | 1999-01-06 | | Release date: | 1999-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase.
Nature, 398, 1999
|
|
7XA9
 
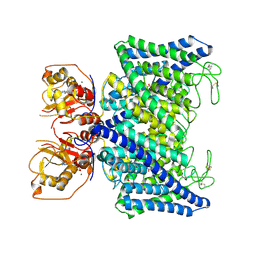 | | Structure of Arabidopsis thaliana CLCa | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chloride channel protein CLC-a, MAGNESIUM ION, ... | | Authors: | Ji, S, Jin, H, Kaiming, Z, Mingxing, W, Shanshan, L, Long, C. | | Deposit date: | 2022-03-17 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of the plant nitrate transporter AtCLCa reveals characteristics of the anion-binding site and the ATP-binding pocket.
J.Biol.Chem., 299, 2023
|
|
3HLN
 
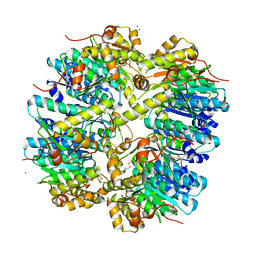 | | Crystal structure of ClpP A153C mutant with inter-heptamer disulfide bonds | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION | | Authors: | Kimber, M.S, Yu, A.Y.H, Borg, M, Chan, H.S, Houry, W.A. | | Deposit date: | 2009-05-27 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Theoretical Studies Indicate that the Cylindrical Protease ClpP Samples Extended and Compact Conformations.
Structure, 18, 2010
|
|
6JYY
 
 | | Crystal structure of the 5-(Hydroxyethyl)-methylthiazole Kinase ThiM from Klebsiella pneumonia | | Descriptor: | Hydroxyethylthiazole kinase | | Authors: | Chen, Y, Wang, L, Shang, F, Lan, J, Liu, W, Xu, Y. | | Deposit date: | 2019-04-29 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight of the 5-(Hydroxyethyl)-methylthiazole kinase ThiM involving vitamin B1 biosynthetic pathway from the Klebsiella pneumoniae.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
3I4B
 
 | | Crystal structure of GSK3b in complex with a pyrimidylpyrrole inhibitor | | Descriptor: | Glycogen synthase kinase-3 beta, N-[(1S)-2-hydroxy-1-phenylethyl]-4-[5-methyl-2-(phenylamino)pyrimidin-4-yl]-1H-pyrrole-2-carboxamide | | Authors: | Ter Haar, E. | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of potent and selective pyrimidylpyrrole inhibitors of extracellular signal-regulated kinase (ERK) using conformational control.
J.Med.Chem., 52, 2009
|
|
6JZE
 
 | | Crystal structure of VASH2-SVBP complex with the magic triangle I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 2 | | Authors: | Chen, Z, Ling, Y, Zeyuan, G, Zhu, L. | | Deposit date: | 2019-05-01 | | Release date: | 2019-08-07 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis of tubulin detyrosination by VASH2/SVBP heterodimer.
Nat Commun, 10, 2019
|
|
1GK0
 
 | | Structure-based prediction of modifications in glutarylamidase to allow single-step enzymatic production of 7-aminocephalosporanic acid from cephalosporin C | | Descriptor: | 1,2-ETHANEDIOL, CEPHALOSPORIN ACYLASE, PHOSPHATE ION | | Authors: | Fritz-Wolf, K, Koller, K.P, Lange, G, Liesum, A, Sauber, K, Schreuder, H, Aretz, W, Kabsch, W. | | Deposit date: | 2001-08-07 | | Release date: | 2002-01-01 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Prediction of Modifications in Glutarylamidase to Allow Single-Step Enzymatic Production of 7-Aminocephalosporanic Acid from Cephalosporin C.
Protein Sci., 11, 2002
|
|
