6HCW
 
 | | Crystal Structure of Lysyl-tRNA Synthetase from Cryptosporidium parvum complexed with L-lysine and a difluoro cyclohexyl chromone ligand | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Robinson, D.A, Baragana, B, Norcross, N, Forte, B, Walpole, C, Gilbert, I.H. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5JHU
 
 | | Potent, Reversible MetAP2 Inhibitors via FBDD | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Dougan, D.R. | | Deposit date: | 2016-04-21 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of potent, reversible MetAP2 inhibitors via fragment based drug discovery and structure based drug design-Part 1.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6S17
 
 | | Crystal Structure of DYRK1A with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ... | | Authors: | Sorrell, F.J, Henderson, S.H, Redondo, C, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Elkins, J.M. | | Deposit date: | 2019-06-18 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Kinase Scaffold Repurposing in the Public Domain
To be published
|
|
7FWW
 
 | | Crystal Structure of human FABP4 in complex with (3-bromophenyl)-[(2Z)-2-phenylimino-1,3-thiazepan-3-yl]methanone | | Descriptor: | (3-bromophenyl)[(2Z)-2-(phenylimino)-1,3-thiazepan-3-yl]methanone, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
9J9D
 
 | | Crystal structure of ALK5 kinase domain in complex with inhibitor HM-279 | | Descriptor: | 1,2-ETHANEDIOL, 2-((1-(but-2-yn-1-yl)-1H-pyrazol-4-yl)(cyclopropylmethyl)amino)-5-(4-(dimethylcarbamoyl)-1H-pyrrol-2-yl)thiazole-4-carboxamide, TGF-beta receptor type-1 | | Authors: | Arai, M, Hanada, M, Taniguchi, H, Ohmoto, H, Naka, K, Sawa, M. | | Deposit date: | 2024-08-22 | | Release date: | 2025-04-02 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (1.337 Å) | | Cite: | Discovery of HM-279, a Potent Inhibitor of ALK5 for Improving Therapeutic Efficacy of Cancer Immunotherapy.
J.Med.Chem., 68, 2025
|
|
5BPY
 
 | |
5ZAP
 
 | | Atomic structure of the herpes simplex virus type 2 B-capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Yuan, S, Wang, J.L, Zhu, D.J, Wang, N, Gao, Q, Chen, W.Y, Tang, H, Wang, J.Z, Zhang, X.Z, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-02-08 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of a herpesvirus capsid at 3.1 angstrom.
Science, 360, 2018
|
|
6ZME
 
 | | SARS-CoV-2 Nsp1 bound to the human CCDC124-80S-eERF1 ribosome complex | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2020-07-02 | | Release date: | 2020-08-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6FND
 
 | | Crystal structure of Toxoplasma gondii AKMT | | Descriptor: | 1,2-ETHANEDIOL, Apical complex lysine methyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pivovarova, Y, Dong, G. | | Deposit date: | 2018-02-02 | | Release date: | 2018-11-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Structure of a Novel Dimeric SET Domain Methyltransferase that Regulates Cell Motility.
J. Mol. Biol., 430, 2018
|
|
6TVL
 
 | | Hen Egg White Lysozyme in complex with a "half sandwich"-type Ru(II) coordination compound | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Chiniadis, L, Giastas, P, Bratsos, I, Papakyriakou, A. | | Deposit date: | 2020-01-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | High-resolution crystal structures of a "half sandwich"-type Ru(II) coordination compound bound to hen egg-white lysozyme and proteinase K.
J.Biol.Inorg.Chem., 25, 2020
|
|
6FPZ
 
 | | Inter-alpha-inhibitor heavy chain 1, D298A | | Descriptor: | ACETATE ION, GLYCEROL, Inter-alpha-trypsin inhibitor heavy chain H1 | | Authors: | Briggs, D.C, Day, A.J. | | Deposit date: | 2018-02-12 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inter-alpha-inhibitor heavy chain-1 has an integrin-like 3D structure mediating immune regulatory activities and matrix stabilization during ovulation
J.Biol.Chem., 2020
|
|
6S5U
 
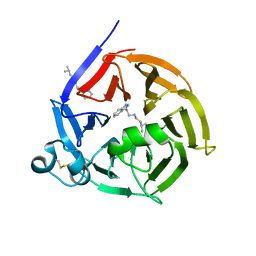 | | Strictosidine Synthase from Ophiorrhiza pumila in complex with N-[2-(1H-Indol-3-yl)ethyl]-3-methyl-1-butanamine | | Descriptor: | Strictosidine synthase, ~{N}-[2-(1~{H}-indol-3-yl)ethyl]-3-methyl-butan-1-amine | | Authors: | Eger, E, Sharma, M, Kroutil, W, Grogan, G. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Inverted Binding of Non-natural Substrates in Strictosidine Synthase Leads to a Switch of Stereochemical Outcome in Enzyme-Catalyzed Pictet-Spengler Reactions.
J.Am.Chem.Soc., 142, 2020
|
|
8UGT
 
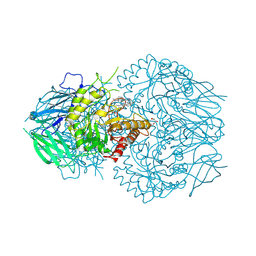 | | E. eligens beta-glucuronidase bound to UNC10206581-G | | Descriptor: | 8-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-5-(methylamino)-1,2,3,4-tetrahydro[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinoline, Beta-glucuronidase, GLYCEROL, ... | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-10-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advanced piperazine-containing inhibitors target microbial beta-glucuronidases linked to gut toxicity.
Rsc Chem Biol, 5, 2024
|
|
6QMC
 
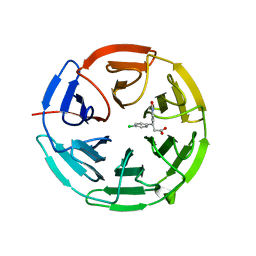 | | Small molecule inhibitor of the KEAP1-NRF2 protein-protein interaction | | Descriptor: | (3~{S})-3-(4-chlorophenyl)-3-(2-oxidanylidene-1~{H}-pyridin-4-yl)propanoic acid, Kelch-like ECH-associated protein 1 | | Authors: | Davies, T.G. | | Deposit date: | 2019-02-07 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure-Activity and Structure-Conformation Relationships of Aryl Propionic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1/Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1/NRF2) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
7FZC
 
 | | Crystal Structure of human FABP4 in complex with 5-[(2-chlorophenoxy)methyl]-4-propan-2-yl-1,2,4-triazole-3-thiol, i.e. SMILES N1(C(=NN=C1COc1c(Cl)cccc1)S)C(C)C with IC50=0.249 microM | | Descriptor: | 5-[(2-chlorophenoxy)methyl]-4-(propan-2-yl)-4H-1,2,4-triazole-3-thiol, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6QMO
 
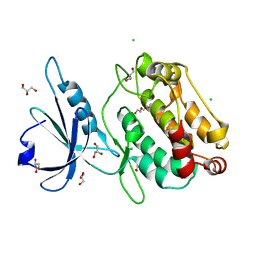 | |
8JST
 
 | |
6TX4
 
 | |
6TXG
 
 | | Proteinase K in complex with a "half sandwich"-type Ru(II) coordination compound | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, NITRATE ION, ... | | Authors: | Chiniadis, L, Giastas, P, Bratsos, I, Papakyriakou, A. | | Deposit date: | 2020-01-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.372 Å) | | Cite: | High-resolution crystal structures of a "half sandwich"-type Ru(II) coordination compound bound to hen egg-white lysozyme and proteinase K.
J.Biol.Inorg.Chem., 25, 2020
|
|
7TY3
 
 | | Crystal Structure of SETD2 Bound to an Indole-based Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETD2, N-[(1R,3R)-3-(4-acetylpiperazin-1-yl)cyclohexyl]-4-fluoro-7-methyl-1H-indole-2-carboxamide, ... | | Authors: | Farrow, N.A. | | Deposit date: | 2022-02-11 | | Release date: | 2022-08-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational-Design-Driven Discovery of EZM0414: A Selective, Potent SETD2 Inhibitor for Clinical Studies.
Acs Med.Chem.Lett., 13, 2022
|
|
5JWV
 
 | | T4 Lysozyme L99A/M102Q with Ethylbenzene Bound | | Descriptor: | Endolysin, PHENYLETHANE | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
7FW6
 
 | | Crystal Structure of human FABP4 in complex with 2-[(3-chlorophenoxy)methyl]-4-phenoxycyclohexane-1-carboxylic acid, i.e. SMILES O(c1ccccc1)[C@H]1CC[C@H]([C@H](C1)COc1cccc(c1)Cl)C(=O)O with IC50=0.259424 microM | | Descriptor: | (1R,2S,4S)-2-[(3-chlorophenoxy)methyl]-4-phenoxycyclohexane-1-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Buettelmann, B, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5JYP
 
 | | Allosteric inhibition of Kidney Isoform of Glutaminase | | Descriptor: | 2-phenyl-~{N}-[5-[(1~{S},3~{S})-3-[5-(2-phenylethanoylamino)-1,3,4-thiadiazol-2-yl]cyclohexyl]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Ramachandran, S, Sivaraman, J. | | Deposit date: | 2016-05-15 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural basis for exploring the allosteric inhibition of human kidney type glutaminase.
Oncotarget, 7, 2016
|
|
6CJE
 
 | | Crystal Structure of Mnk2-D228G in complex with Inhibitor | | Descriptor: | 4-[(9H-purin-6-yl)amino]benzamide, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-26 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
6UK6
 
 | |
