5URQ
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p176 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-{2-chloro-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]phenyl}-alpha-D-ribofuranosylamine, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-12 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p176
To Be Published
|
|
6QYA
 
 | |
9JQ0
 
 | |
7CFO
 
 | | Crystal structure of human RXRalpha ligand binding domain complexed with CBTF-EE. | | Descriptor: | 1-[3-(2-ethoxyethoxy)-5,5,8,8-tetramethyl-6,7-dihydronaphthalen-2-yl]-2-(trifluoromethyl)benzimidazole-5-carboxylic acid, GLYCEROL, Retinoic acid receptor RXR-alpha | | Authors: | Watanabe, M, Fujihara, M, Motoyama, T, Kawasaki, M, Yamada, S, Takamura, Y, Ito, S, Makishima, M, Nakano, S, Kakuta, H. | | Deposit date: | 2020-06-27 | | Release date: | 2021-01-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a "Gatekeeper" Antagonist that Blocks Entry Pathway to Retinoid X Receptors (RXRs) without Allosteric Ligand Inhibition in Permissive RXR Heterodimers.
J.Med.Chem., 64, 2021
|
|
5NY1
 
 | | Carbonic Anhydrase II Inhibitor RA10 | | Descriptor: | 3,6,7-trimethyl-~{N}-[(4-sulfamoylphenyl)methyl]-1-benzofuran-2-carboxamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
6R2N
 
 | | Crystal structure of KlGlk1 glucokinase from Kluyveromyces lactis | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Glucokinase-1 | | Authors: | Zak, K, Wator, E, Grudnik, P. | | Deposit date: | 2019-03-18 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Crystal Structure of Kluyveromyces lactis Glucokinase ( Kl Glk1).
Int J Mol Sci, 20, 2019
|
|
5O02
 
 | |
9JOC
 
 | |
5YT3
 
 | | Structure of the Human Mitogen-Activated Protein Kinase Kinase 1 S218D and S222D mutant | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase kinase 1, isoform CRA_d, ... | | Authors: | Nakae, S, Doko, K, Tada, T, Shirai, T. | | Deposit date: | 2017-11-16 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Human Mitogen-Activated Protein Kinase Kinase 1 S218D and S222D mutant
To Be Published
|
|
6R2R
 
 | |
6IBX
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 5 | | Descriptor: | 3-[[8-(1-methylindol-6-yl)quinoxalin-6-yl]amino]-~{N}-[(3~{S})-1-methylpiperidin-3-yl]pyridine-4-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, ... | | Authors: | Banaszak, K, Pawlik, H, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-12-01 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Synthesis of amide and sulfonamide substituted N-aryl 6-aminoquinoxalines as PFKFB3 inhibitors with improved physicochemical properties.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
7UPS
 
 | |
9GGD
 
 | |
6C7Y
 
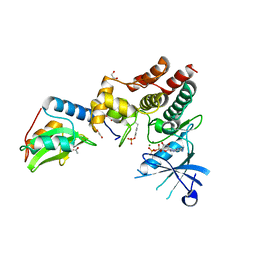 | | Crystal structure of inhibitory protein SOCS1 in complex with JAK1 kinase domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Liau, N.P.D, Laktyushin, A, Lucet, I.S, Murphy, J.M, Yao, S, Callaghan, K, Nicola, N.A, Kershaw, N.J, Babon, J.J. | | Deposit date: | 2018-01-23 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | The molecular basis of JAK/STAT inhibition by SOCS1.
Nat Commun, 9, 2018
|
|
4YLL
 
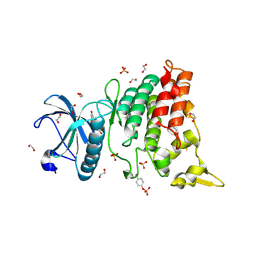 | | Crystal structure of DYRK1AA in complex with 10-Bromo-substituted 11H-indolo[3,2-c]quinolone-6-carboxylic acid inhibitor 5t | | Descriptor: | 1,2-ETHANEDIOL, 10-bromo-2-iodo-11H-indolo[3,2-c]quinoline-6-carboxylic acid, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, ... | | Authors: | Chaikuad, A, Falke, H, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-03-05 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 10-Iodo-11H-indolo[3,2-c]quinoline-6-carboxylic Acids Are Selective Inhibitors of DYRK1A.
J.Med.Chem., 58, 2015
|
|
9GGB
 
 | |
8BF3
 
 | |
4AAF
 
 | | Crystal structure of the mutant D75N I-CreI in complex with an altered target (The four central bases, 2NN region, are composed by TGCA from 5' to 3') | | Descriptor: | 1,2-ETHANEDIOL, 24MER DNA, DNA ENDONUCLEASE I-CREI | | Authors: | Molina, R, Redondo, P, Stella, S, Marenchino, M, D'Abramo, M, Gervasio, F.L, Epinat, J.C, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-Specific Protein-DNA Interactions Control I-Crei Target Binding and Cleavage.
Nucleic Acids Res., 40, 2012
|
|
8EJB
 
 | |
5O52
 
 | | Glycogen Phosphorylase b in complex with 33b | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{R})-5-azanyl-2-(hydroxymethyl)-6-(4-naphthalen-2-yl-1~{H}-imidazol-2-yl)oxane-3,4-diol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kantsadi, A.L, Kyriakis, E, Stravodimos, G.A, Solovou, T.G.A, Chatzileontiadou, D.S, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2017-05-31 | | Release date: | 2017-09-27 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nanomolar Inhibitors of Glycogen Phosphorylase Based on beta-d-Glucosaminyl Heterocycles: A Combined Synthetic, Enzyme Kinetic, and Protein Crystallography Study.
J. Med. Chem., 60, 2017
|
|
8SWA
 
 | | Crystal structure of the human S-adenosylmethionine synthetase 1 in complex with SAM and PPNP | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Bruemmer, K.J, Pham, V.N, Toh, J.D.W. | | Deposit date: | 2023-05-18 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Formaldehyde regulates S -adenosylmethionine biosynthesis and one-carbon metabolism.
Science, 382, 2023
|
|
6FD5
 
 | | Crystal Structure of Human APRT-Tyr105Phe variant in complex with Adenine, PRPP and Mg2+, 14 days post crystallization (with AMP and PPi products partially generated) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, ADENOSINE MONOPHOSPHATE, Adenine phosphoribosyltransferase, ... | | Authors: | Nioche, P, Huyet, J, Ozeir, M. | | Deposit date: | 2017-12-21 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insights into the Forward and Reverse Enzymatic Reactions in Human Adenine Phosphoribosyltransferase.
Cell Chem Biol, 25, 2018
|
|
7V35
 
 | | Cryo-EM structure of the GIPR/GLP-1R/GCGR triagonist peptide 20-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, F.h, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-10 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
5NV7
 
 | |
5JAH
 
 | |
