4E6E
 
 | |
4U39
 
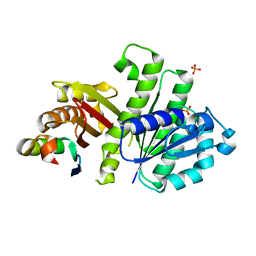 | | Crystal Structure of FtsZ:MciZ Complex from Bacillus subtilis | | Descriptor: | Cell division factor, Cell division protein FtsZ, PHOSPHATE ION | | Authors: | Bisson-Filho, A.W, Discola, K.F, Castellen, P, Blasios, V, Martins, A, Sforca, M.L, Garcia, W, Zeri, A.C, Erickson, H.P, Dessen, A, Gueiros-Filho, F.J. | | Deposit date: | 2014-07-19 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Crystal Structure of FtsZ:MciZ Complex from Bacillus subtilis
To be Published
|
|
1FSZ
 
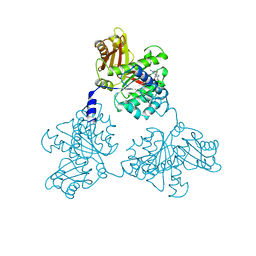 | |
4DXD
 
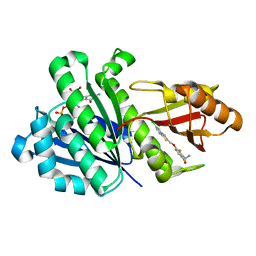 | | Staphylococcal Aureus FtsZ in complex with 723 | | Descriptor: | 3-[(6-chloro[1,3]thiazolo[5,4-b]pyridin-2-yl)methoxy]-2,6-difluorobenzamide, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Lu, J, Soisson, S.M. | | Deposit date: | 2012-02-27 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Restoring methicillin-resistant Staphylococcus aureus susceptibility to beta-lactam antibiotics.
Sci Transl Med, 4, 2012
|
|
8IBN
 
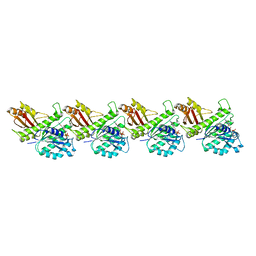 | | Cryo-EM structure of KpFtsZ single filament | | Descriptor: | Cell division protein FtsZ, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, POTASSIUM ION | | Authors: | Fujita, J, Amesaka, H, Yoshizawa, T, Kuroda, N, Kamimura, N, Hibino, K, Konishi, T, Kato, Y, Hara, M, Inoue, T, Namba, K, Tanaka, S, Matsumura, H. | | Deposit date: | 2023-02-10 | | Release date: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody
Nat Commun, 14, 2023
|
|
8HTB
 
 | | Staphylococcus aureus FtsZ 12-316 complexed with TXH9179 | | Descriptor: | 3-[(6-ethynyl-[1,3]thiazolo[5,4-b]pyridin-2-yl)methoxy]-2,6-bis(fluoranyl)benzamide, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Bryan, E, Ferrer-Gonzalez, E, Sagong, H.Y, Fujita, J, Mark, L, Kaul, M, LaVoie, E.J, Matsumura, H, Pilch, D.S. | | Deposit date: | 2022-12-21 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and Antibacterial Characterization of a New Benzamide FtsZ Inhibitor with Superior Bactericidal Activity and In Vivo Efficacy Against Multidrug-Resistant Staphylococcus aureus.
Acs Chem.Biol., 18, 2023
|
|
6UMK
 
 | | Structure of E. coli FtsZ(L178E)-GDP complex | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-10-09 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High-resolution crystal structures of Escherichia coli FtsZ bound to GDP and GTP.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4KWE
 
 | | GDP-bound, double-stranded, curved FtsZ protofilament structure | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Li, Y, Hsin, J, Zhao, L, Cheng, Y, Shang, W, Huang, K.C, Wang, H.W, Ye, S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation
Science, 341, 2013
|
|
6UNX
 
 | | Structure of E. coli FtsZ(L178E)-GTP complex | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-10-13 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution crystal structures of Escherichia coli FtsZ bound to GDP and GTP.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4M8I
 
 | | 1.43 Angstrom resolution crystal structure of cell division protein FtsZ (ftsZ) from Staphylococcus epidermidis RP62A in complex with GDP | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Halavaty, A.S, Minasov, G, Winsor, J, Dubrovska, I, Filippova, E.V, Olsen, D.B, Therien, A, Shuvalova, L, Young, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-13 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | 1.43 Angstrom resolution crystal structure of cell division protein FtsZ (ftsZ) from Staphylococcus epidermidis RP62A in complex with GDP
To be Published
|
|
7YOP
 
 | | Spiroplasma melliferum FtsZ bound to GMPPNP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chakraborty, J, Pananghat, G. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.20003128 Å) | | Cite: | Structural basis of kinetic polarity of FtsZ from a cell wall-less bacterium Spiroplasma
To Be Published
|
|
7YSZ
 
 | | Spiroplasma melliferum FtsZ bound to GDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Chakraborty, J, Pananghat, G. | | Deposit date: | 2022-08-13 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3000412 Å) | | Cite: | Structural basis of kinetic polarity of FtsZ from a cell wall less bacterium Spiroplasma
To Be Published
|
|
5XDT
 
 | | Staphylococcus aureus FtsZ 12-316 complexed with TXA707 | | Descriptor: | 1-methylpyrrolidin-2-one, 2,6-bis(fluoranyl)-3-[[6-(trifluoromethyl)-[1,3]thiazolo[5,4-b]pyridin-2-yl]methoxy]benzamide, CALCIUM ION, ... | | Authors: | Fujita, J, Maeda, Y, Mizohata, E, Inoue, T, Kaul, M, Parhi, A.K, LaVoie, E.J, Pilch, D.S, Matsumura, H. | | Deposit date: | 2017-03-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Flexibility of an Inhibitor Overcomes Drug Resistance Mutations in Staphylococcus aureus FtsZ
ACS Chem. Biol., 12, 2017
|
|
5XDW
 
 | | Staphylococcus aureus FtsZ 12-316 G196S | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Maeda, Y, Mizohata, E, Inoue, T, Kaul, M, Parhi, A.K, LaVoie, E.J, Pilch, D.S, Matsumura, H. | | Deposit date: | 2017-03-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Flexibility of an Inhibitor Overcomes Drug Resistance Mutations in Staphylococcus aureus FtsZ
ACS Chem. Biol., 12, 2017
|
|
5H5I
 
 | | Staphylococcus aureus FtsZ-GDP R29A mutant in R state | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
5H5H
 
 | | Staphylococcus aureus FtsZ-GDP R29A mutant in T state | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
5H5G
 
 | | Staphylococcus aureus FtsZ-GDP in T and R states | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
5ZUE
 
 | | GTP-bound, double-stranded, curved FtsZ protofilament structure | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Guan, F, Yu, J, Yu, J, Liu, Y, Li, Y, Feng, X.H, Huang, K.C, Chang, Z, Ye, S. | | Deposit date: | 2018-05-07 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Lateral interactions between protofilaments of the bacterial tubulin homolog FtsZ are essential for cell division
Elife, 7, 2018
|
|
5V68
 
 | | Crystal structure of cell division protein FtsZ from Mycobacterium tuberculosis bounded via the T9 loop | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Lazo, E.O, Ojima, I, Chowdhury, S.R, Awasthi, D, Jakoncic, J. | | Deposit date: | 2017-03-16 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Novel T9 loop conformation of filamenting temperature-sensitive mutant Z from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5XDU
 
 | | Staphylococcus aureus FtsZ 12-316 complexed with TXA6101 | | Descriptor: | 3-[[5-bromanyl-4-[4-(trifluoromethyl)phenyl]-1,3-oxazol-2-yl]methoxy]-2,6-bis(fluoranyl)benzamide, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Fujita, J, Maeda, Y, Mizohata, E, Inoue, T, Kaul, M, Parhi, A.K, LaVoie, E.J, Pilch, D.S, Matsumura, H. | | Deposit date: | 2017-03-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Flexibility of an Inhibitor Overcomes Drug Resistance Mutations in Staphylococcus aureus FtsZ
ACS Chem. Biol., 12, 2017
|
|
5XDV
 
 | | Staphylococcus aureus FtsZ 12-316 G196S complexed with TXA6101 | | Descriptor: | 3-[[5-bromanyl-4-[4-(trifluoromethyl)phenyl]-1,3-oxazol-2-yl]methoxy]-2,6-bis(fluoranyl)benzamide, CALCIUM ION, Cell division protein FtsZ, ... | | Authors: | Fujita, J, Maeda, Y, Mizohata, E, Inoue, T, Kaul, M, Parhi, A.K, LaVoie, E.J, Pilch, D.S, Matsumura, H. | | Deposit date: | 2017-03-30 | | Release date: | 2017-08-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Flexibility of an Inhibitor Overcomes Drug Resistance Mutations in Staphylococcus aureus FtsZ
ACS Chem. Biol., 12, 2017
|
|
2VAW
 
 | | FtsZ Pseudomonas aeruginosa GDP | | Descriptor: | CELL DIVISION PROTEIN FTSZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Oliva, M.A, Lowe, J. | | Deposit date: | 2007-09-04 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insights Into the Conformational Variability of Ftsz
J.Mol.Biol., 373, 2007
|
|
2VAP
 
 | | FtsZ GDP M. jannaschii | | Descriptor: | CELL DIVISION PROTEIN FTSZ HOMOLOG 1, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Oliva, M.A, Lowe, J. | | Deposit date: | 2007-09-03 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights Into the Conformational Variability of Ftsz
J.Mol.Biol., 373, 2007
|
|
2VAM
 
 | | FtsZ B. subtilis | | Descriptor: | CELL DIVISION PROTEIN FTSZ, SULFATE ION | | Authors: | Oliva, M.A, Lowe, J. | | Deposit date: | 2007-09-03 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights Into the Conformational Variability of Ftsz
J.Mol.Biol., 373, 2007
|
|
2VXY
 
 | | The structure of FTsZ from Bacillus subtilis at 1.7A resolution | | Descriptor: | CELL DIVISION PROTEIN FTSZ, CITRIC ACID, POTASSIUM ION | | Authors: | Barynin, V.V, Baker, P.J, Rice, D.W, Sedelnikova, S.E, Haydon, D.J, Stokes, N.R, Ure, R, Galbraith, G, Bennett, J.M, Brown, D.R, Heal, J.R, Sheridan, J.M, Aiwale, S.T, Chauhan, P.K, Srivastava, A, Taneja, A, Collins, I, Errington, J, Czaplewski, L.G. | | Deposit date: | 2008-07-15 | | Release date: | 2008-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An Inhibitor of Ftsz with Potent and Selective Anti-Staphylococcal Activity.
Science, 321, 2008
|
|
