1I9Y
 
 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 5-PHOSPHATASE DOMAIN (IPP5C) OF SPSYNAPTOJANIN | | Descriptor: | PHOSPHATIDYLINOSITOL PHOSPHATE PHOSPHATASE | | Authors: | Tsujishita, Y, Guo, S, Stolz, L, York, J.D, Hurley, J.H. | | Deposit date: | 2001-03-21 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate 5-phosphatase.
Cell(Cambridge,Mass.), 105, 2001
|
|
3K55
 
 | | Structure of beta hairpin deletion mutant of beta toxin from Staphylococcus aureus | | Descriptor: | Beta-hemolysin, CHLORIDE ION, SODIUM ION | | Authors: | Kruse, A.C, Huseby, M, Shi, K, Digre, J, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2009-10-06 | | Release date: | 2011-01-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structure of a mutant beta toxin from Staphylococcus aureus reveals domain swapping and conformational flexibility
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3TEB
 
 | | endonuclease/exonuclease/phosphatase family protein from Leptotrichia buccalis C-1013-b | | Descriptor: | Endonuclease/exonuclease/phosphatase, MAGNESIUM ION | | Authors: | Chang, C, Bigelow, L, Muniez, I, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-08-12 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of endonuclease/exonuclease/phosphatase family protein from Leptotrichia buccalis C-1013-b
To be Published
|
|
1VYB
 
 | | Endonuclease domain of human LINE1 ORF2p | | Descriptor: | GLYCEROL, ORF2 CONTAINS A REVERSE TRANSCRIPTASE DOMAIN, SULFATE ION, ... | | Authors: | Weichenrieder, O, Repanas, K, Perrakis, A. | | Deposit date: | 2004-04-25 | | Release date: | 2004-06-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the targeting endonuclease of the human LINE-1 retrotransposon.
Structure, 12, 2004
|
|
1I9Z
 
 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 5-PHOSPHATASE DOMAIN (IPP5C) OF SPSYNAPTOJANIN IN COMPLEX WITH INOSITOL (1,4)-BISPHOSPHATE AND CALCIUM ION | | Descriptor: | CALCIUM ION, D-MYO-INOSITOL-1,4-BISPHOSPHATE, PHOSPHATIDYLINOSITOL PHOSPHATE PHOSPHATASE | | Authors: | Tsujishita, Y, Guo, S, Stolz, L, York, J.D, Hurley, J.H. | | Deposit date: | 2001-03-21 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate 5-phosphatase.
Cell(Cambridge,Mass.), 105, 2001
|
|
6LPM
 
 | |
2JC5
 
 | | Apurinic Apyrimidinic (AP) endonuclease (NApe) from Neisseria Meningitidis | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, BICINE, EXODEOXYRIBONUCLEASE, ... | | Authors: | Carpenter, E.P, Corbett, A, Thomson, H, Adacha, J, Jensen, K, Bergeron, J, Kasampalidis, I, Exley, R, Winterbotham, M, Tang, C, Baldwin, G.S, Freemont, P. | | Deposit date: | 2006-12-19 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ap Endonuclease Paralogues with Distinct Activities in DNA Repair and Bacterial Pathogenesis.
Embo J., 26, 2007
|
|
2JC4
 
 | | 3'-5' exonuclease (NExo) from Neisseria Meningitidis | | Descriptor: | ACETATE ION, DIHYDROGENPHOSPHATE ION, EXODEOXYRIBONUCLEASE III, ... | | Authors: | Carpenter, E.P, Corbett, A, Thomson, H, Adacha, J, Jensen, K, Bergeron, J, Kasampalidis, I, Exley, R, Winterbotham, M, Tang, C, Baldwin, G, Freemont, P. | | Deposit date: | 2006-12-19 | | Release date: | 2007-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ap Endonuclease Paralogues with Distinct Activities in DNA Repair and Bacterial Pathogenesis.
Embo J., 26, 2007
|
|
7SUV
 
 | | APE1 exonuclease substrate complex with 8oxoG opposite A | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Whitaker, A.W, Freudenthal, B.D. | | Deposit date: | 2021-11-18 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Processing oxidatively damaged bases at DNA strand breaks by APE1.
Nucleic Acids Res., 50, 2022
|
|
7SVB
 
 | | APE1 exonuclease substrate complex with 8oxoG opposite C | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(8OG))-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*CP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(P*TP*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Whitaker, A.W, Freudenthal, B.D. | | Deposit date: | 2021-11-18 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Processing oxidatively damaged bases at DNA strand breaks by APE1.
Nucleic Acids Res., 50, 2022
|
|
8IGI
 
 | | Crystal structure of HP1526 (XthA)- a base excision DNA repair protein in Helicobacter pylori | | Descriptor: | 1,3-BUTANEDIOL, Exodeoxyribonuclease (LexA), MANGANESE (II) ION | | Authors: | Dinh, T.T, Dao, O, Lee, K.H. | | Deposit date: | 2023-02-20 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of the apurinic/apyrimidinic endonuclease XthA (HP1526 protein) from Helicobacter pylori.
Biochem.Biophys.Res.Commun., 663, 2023
|
|
2O3H
 
 | | Crystal structure of the human C65A Ape | | Descriptor: | ACETATE ION, DNA-(apurinic or apyrimidinic site) lyase, SAMARIUM (III) ION | | Authors: | Georgiadis, M.M, Gaur, R.K, Delaplane, S, Svenson, J. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease
Mutat.Res., 643, 2008
|
|
2O3C
 
 | | Crystal structure of zebrafish Ape | | Descriptor: | APEX nuclease 1, LEAD (II) ION | | Authors: | Georgiadis, M.M, Gaur, R.K, Delaplane, S, Svenson, J. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease
Mutat.Res., 643, 2008
|
|
8SP5
 
 | |
8SP7
 
 | |
8J2F
 
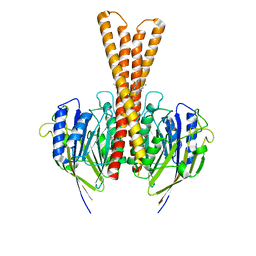 | | Human neutral shpingomyelinase | | Descriptor: | HEPTANE, MAGNESIUM ION, Sphingomyelin phosphodiesterase 2, ... | | Authors: | Zhang, S.S. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Molecular basis for the catalytic mechanism of human neutral sphingomyelinases 1 (hSMPD2).
Nat Commun, 14, 2023
|
|
3DNI
 
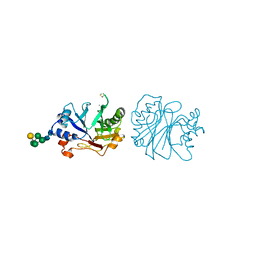 | | CRYSTALLOGRAPHIC REFINEMENT AND STRUCTURE OF DNASE I AT 2 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, DEOXYRIBONUCLEASE I, alpha-D-galactopyranose-(1-6)-beta-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Oefner, C, Suck, D. | | Deposit date: | 1992-08-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic refinement and structure of DNase I at 2 A resolution.
J.Mol.Biol., 192, 1986
|
|
6FK5
 
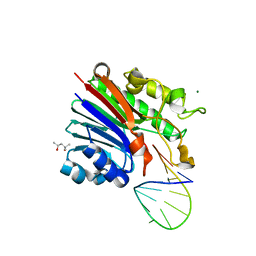 | | Structure of 3' phosphatase NExo (D146N) from Neisseria bound to DNA substrate in presence of magnesium ion | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(P*CP*TP*AP*GP*CP*GP*AP*AP*GP*CP*TP*AP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Silhan, J, Zhao, Q, Boura, E, Thomson, H, Foster, A, Tang, C.M, Freemont, P.S, Baldwin, G.S. | | Deposit date: | 2018-01-23 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for recognition and repair of the 3'-phosphate by NExo, a base excision DNA repair nuclease from Neisseria meningitidis.
Nucleic Acids Res., 46, 2018
|
|
6P94
 
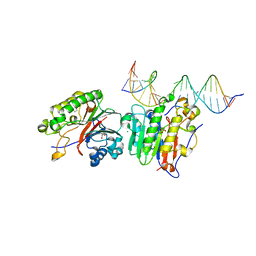 | | Human APE1 C65A AP-endonuclease product complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Whitaker, A.W, Stark, W.J, Freudenthal, B.D. | | Deposit date: | 2019-06-09 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Functions of the major abasic endonuclease (APE1) in cell viability and genotoxin resistance.
Mutagenesis, 35, 2020
|
|
6P93
 
 | | Human APE1 K98A AP-endonuclease product complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Whitaker, A.W, Stark, W.J, Freudenthal, B.D. | | Deposit date: | 2019-06-09 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functions of the major abasic endonuclease (APE1) in cell viability and genotoxin resistance.
Mutagenesis, 35, 2020
|
|
6FKE
 
 | | Structure of 3' phosphatase NExo (D146N) from Neisseria bound to product DNA hairpin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*TP*AP*GP*CP*GP*AP*AP*GP*CP*TP*A)-3'), Exodeoxyribonuclease III, ... | | Authors: | Silhan, J, Zhao, Q, Boura, E, Thomson, H, Foster, A, Tang, C.M, Freemont, P.S, Baldwin, G.S. | | Deposit date: | 2018-01-23 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structural basis for recognition and repair of the 3'-phosphate by NExo, a base excision DNA repair nuclease from Neisseria meningitidis.
Nucleic Acids Res., 46, 2018
|
|
6FK4
 
 | | Structure of 3' phosphatase NExo (WT) from Neisseria bound to DNA substrate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(P*GP*CP*TP*AP*GP*CP*GP*AP*AP*GP*CP*TP*AP*GP*A)-3'), Exodeoxyribonuclease III | | Authors: | Silhan, J, Zhao, Q, Boura, E, Thomson, H, Foster, A, Tang, C.M, Freemont, P.S, Baldwin, G.S. | | Deposit date: | 2018-01-23 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Structural basis for recognition and repair of the 3'-phosphate by NExo, a base excision DNA repair nuclease from Neisseria meningitidis.
Nucleic Acids Res., 46, 2018
|
|
1AKO
 
 | | EXONUCLEASE III FROM ESCHERICHIA COLI | | Descriptor: | EXONUCLEASE III | | Authors: | Mol, C.D, Kuo, C.-F, Thayer, M.M, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 1997-05-26 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of the multifunctional DNA-repair enzyme exonuclease III.
Nature, 374, 1995
|
|
8KA5
 
 | |
8KA3
 
 | |
