6DQX
 
 | | Actinobacillus ureae class Id ribonucleotide reductase alpha subunit | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | McBride, M.J, Palowitch, G.M, Boal, A.K. | | Deposit date: | 2018-06-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of Class Id Ribonucleotide Reductase Catalytic Subunits Reveal a Minimal Architecture for Deoxynucleotide Biosynthesis.
Biochemistry, 58, 2019
|
|
6DQW
 
 | |
7URG
 
 | |
7B9Q
 
 | | The SERp optimized structure of Ribonucleotide reductase from Rhodobacter sphaeroides | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, Vitamin B12-dependent ribonucleotide reductase | | Authors: | Loderer, C, Feiler, C, Wilk, P, Kabinger, F. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | HUG Domain Is Responsible for Active Dimer Stabilization in an NrdJd Ribonucleotide Reductase.
Biochemistry, 61, 2022
|
|
7B9P
 
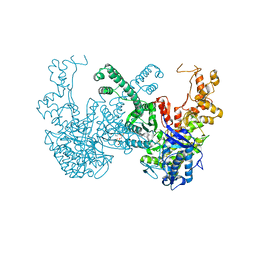 | | Structure of Ribonucleotide reductase from Rhodobacter sphaeroides | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, Vitamin B12-dependent ribonucleotide reductase | | Authors: | Wilk, P, Feiler, C, Loderer, C, Kabinger, F. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.646 Å) | | Cite: | HUG Domain Is Responsible for Active Dimer Stabilization in an NrdJd Ribonucleotide Reductase.
Biochemistry, 61, 2022
|
|
2R1R
 
 | |
6W4X
 
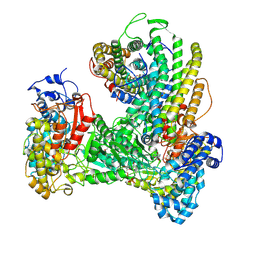 | | Holocomplex of E. coli class Ia ribonucleotide reductase with GDP and TTP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MU-OXO-DIIRON, ... | | Authors: | Kang, G, Taguchi, A, Stubbe, J, Drennan, C.L. | | Deposit date: | 2020-03-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a trapped radical transfer pathway within a ribonucleotide reductase holocomplex.
Science, 368, 2020
|
|
5TUS
 
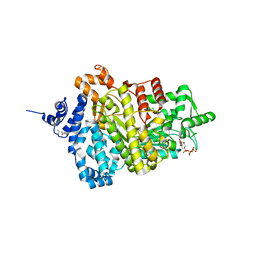 | | Potent competitive inhibition of human ribonucleotide reductase by a novel non-nucleoside small molecule | | Descriptor: | 2-hydroxy-N'-[(Z)-(2-hydroxynaphthalen-1-yl)methylidene]benzohydrazide, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | Authors: | Mohammed, F.A, Alam, I, Dealwis, C.G. | | Deposit date: | 2016-11-07 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Potent competitive inhibition of human ribonucleotide reductase by a nonnucleoside small molecule.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4X3V
 
 | | Crystal structure of human ribonucleotide reductase 1 bound to inhibitor | | Descriptor: | N~6~-{N-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)acetyl]-2-methyl-D-alanyl}-D-lysine, Ribonucleoside-diphosphate reductase large subunit, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Dealwis, C.G, Ahmad, M.F, Alam, I. | | Deposit date: | 2014-12-01 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Identification of Non-nucleoside Human Ribonucleotide Reductase Modulators.
J.Med.Chem., 58, 2015
|
|
7R1R
 
 | | RIBONUCLEOTIDE REDUCTASE E441Q MUTANT R1 PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | RIBONUCLEOTIDE REDUCTASE R1 PROTEIN, RIBONUCLEOTIDE REDUCTASE R2 PROTEIN | | Authors: | Eriksson, M, Eklund, H. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase.
J.Biol.Chem., 272, 1997
|
|
4R1R
 
 | |
6R1R
 
 | | RIBONUCLEOTIDE REDUCTASE E441D MUTANT R1 PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | RIBONUCLEOTIDE REDUCTASE R1 PROTEIN, RIBONUCLEOTIDE REDUCTASE R2 PROTEIN | | Authors: | Eriksson, M, Eklund, H. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase.
J.Biol.Chem., 272, 1997
|
|
7MDI
 
 | | Structure of the Neisseria gonorrhoeae ribonucleotide reductase in the inactive state | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Levitz, T.S, Drennan, C.L, Brignole, E.J. | | Deposit date: | 2021-04-05 | | Release date: | 2022-01-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Effects of chameleon dispense-to-plunge speed on particle concentration, complex formation, and final resolution: A case study using the Neisseria gonorrhoeae ribonucleotide reductase inactive complex.
J.Struct.Biol., 214, 2021
|
|
5IM3
 
 | |
5R1R
 
 | | RIBONUCLEOTIDE REDUCTASE E441A MUTANT R1 PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | RIBONUCLEOTIDE REDUCTASE R1 PROTEIN, RIBONUCLEOTIDE REDUCTASE R2 PROTEIN | | Authors: | Eriksson, M, Eklund, H. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase.
J.Biol.Chem., 272, 1997
|
|
7RW6
 
 | | BORF2-APOBEC3Bctd Complex | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3B, Maltose/maltodextrin-binding periplasmic protein,Ribonucleoside-diphosphate reductase large subunit, ZINC ION | | Authors: | Shaban, N.M, Yan, R, Shi, K, McLellan, J.S, Yu, Z, Harris, R.S. | | Deposit date: | 2021-08-19 | | Release date: | 2022-04-27 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM structure of the EBV ribonucleotide reductase BORF2 and mechanism of APOBEC3B inhibition.
Sci Adv, 8, 2022
|
|
6CGL
 
 | |
6CGN
 
 | |
4ERP
 
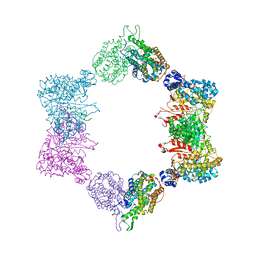 | |
6AUI
 
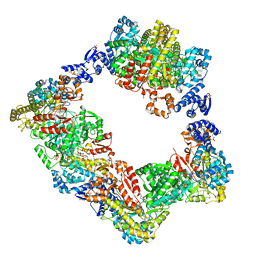 | | Human ribonucleotide reductase large subunit (alpha) with dATP and CDP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Brignole, E.J, Drennan, C.L, Asturias, F.J, Tsai, K.L, Penczek, P.A. | | Deposit date: | 2017-09-01 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | 3.3- angstrom resolution cryo-EM structure of human ribonucleotide reductase with substrate and allosteric regulators bound.
Elife, 7, 2018
|
|
6CGM
 
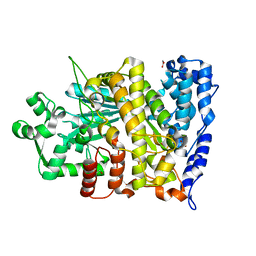 | |
4ERM
 
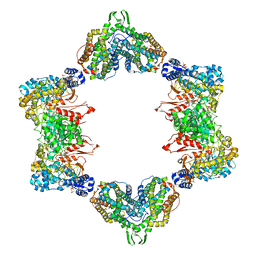 | |
3S8A
 
 | | Structure of Yeast Ribonucleotide Reductase R293A with dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large chain 1 | | Authors: | Ahmad, M.F, Kaushal, P.S, Wan, Q, Wijeratna, S.R, Huang, M, Dealwis, C.D. | | Deposit date: | 2011-05-27 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and biochemical basis of lethal mutant R293A of yeast ribonucleotide reductase
To be Published
|
|
3TBA
 
 | | Structure of Yeast Ribonucleotide Reductase 1 Q288A with dGTP and ADP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ahmad, M.F, Kaushal, P.S, Wan, Q, Wijeratna, S.R, Huang, M, Dealwis, C. | | Deposit date: | 2011-08-05 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Role of Arginine 293 and Glutamine 288 in Communication between Catalytic and Allosteric Sites in Yeast Ribonucleotide Reductase.
J.Mol.Biol., 419, 2012
|
|
3TB9
 
 | | Structure of Yeast Ribonucleotide Reductase 1 Q288A with AMPPNP and CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Ahmad, M.F, Kaushal, P.S, Wan, Q, Wijeratna, S.R, Huang, M, Dealwis, C.D. | | Deposit date: | 2011-08-05 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Role of Arginine 293 and Glutamine 288 in Communication between Catalytic and Allosteric Sites in Yeast Ribonucleotide Reductase.
J.Mol.Biol., 419, 2012
|
|
