8OTK
 
 | | Structure of ClpC Q11P N-terminal Domain | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC / Negative regulator of tic competence clcC/mecB, CARBONATE ION, GLYCEROL, ... | | Authors: | Evans, N.J, Isaacson, R.L, Camp, A.H. | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A novel ClpC-ClpP adaptor protein that functions in the developing Bacillus subtilis spore
To Be Published
|
|
4P15
 
 | | Structure of the ClpC N-terminal domain from an alkaliphilic Bacillus lehensis G1 species | | Descriptor: | Bacillus lehensis ClpC N-terminal domain, SULFATE ION | | Authors: | Rashid, S.A, Littler, D.R, Illias, R.M, Murad, A.M.A, Rossjohn, J, Beddoe, T, Mahadi, N.M. | | Deposit date: | 2014-01-31 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the ClpC N-terminal domain at 1.85 Angstroms resolution from an alkaliphilic Bacillus lehensis G1 species
To Be Published
|
|
7TFM
 
 | | Atomic Structure of the Leishmania spp. Hsp100 N-Domain | | Descriptor: | ATP-dependent Clp protease subunit, heat shock protein 100 (HSP100), GLYCEROL | | Authors: | Mercado, J.M, Lee, S, Chang, C, Sung, N, Soong, L, Catic, A, Tsai, F.T.F. | | Deposit date: | 2022-01-06 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.055 Å) | | Cite: | Atomic structure of the Leishmania spp. Hsp100 N-domain.
Proteins, 90, 2022
|
|
9DIN
 
 | |
2K77
 
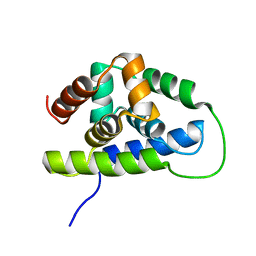 | | NMR solution structure of the Bacillus subtilis ClpC N-domain | | Descriptor: | Negative regulator of genetic competence clpC/mecB | | Authors: | Kojetin, D.J, McLaughlin, P.D, Thompson, R.J, Rance, M, Cavanagh, J. | | Deposit date: | 2008-08-04 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural and motional contributions of the Bacillus subtilis ClpC N-domain to adaptor protein interactions.
J.Mol.Biol., 387, 2009
|
|
8B9O
 
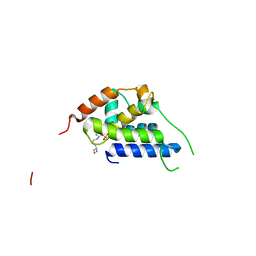 | |
8B9U
 
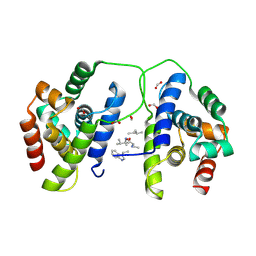 | |
7AA4
 
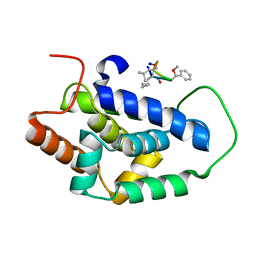 | | Structure of ClpC1-NTD bound to a CymA analogue | | Descriptor: | Negative regulator of genetic competence ClpC/mecB, polymer Cyclomarin A analogue | | Authors: | Meinhart, A, Morreale, F.E, Kaiser, M, Clausen, T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
8IBO
 
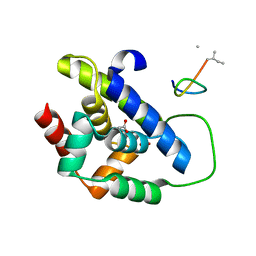 | |
8IBP
 
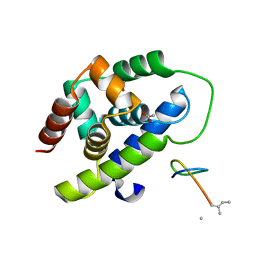 | |
4IRF
 
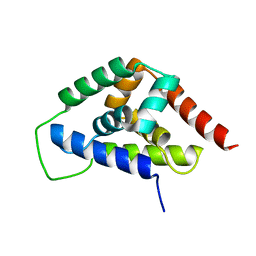 | |
2Y1Q
 
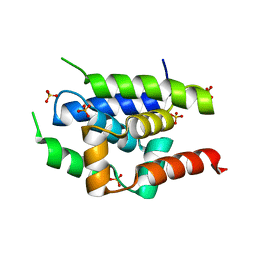 | | Crystal Structure of ClpC N-terminal Domain | | Descriptor: | NEGATIVE REGULATOR OF GENETIC COMPETENCE CLPC/MECB, SULFATE ION | | Authors: | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Mechanism of the Hexameric Meca-Clpc Molecular Machine.
Nature, 471, 2011
|
|
3FES
 
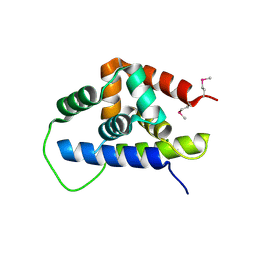 | | Crystal Structure of the ATP-dependent Clp Protease ClpC from Clostridium difficile | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ATP-dependent Clp endopeptidase, MAGNESIUM ION, ... | | Authors: | Kim, Y, Tesar, C, Li, H, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-01 | | Release date: | 2008-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structure of the ATP-dependent Clp Protease ClpC from Clostridium difficile
To be Published
|
|
4IOD
 
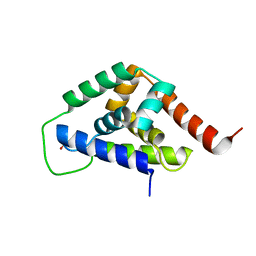 | |
4HH6
 
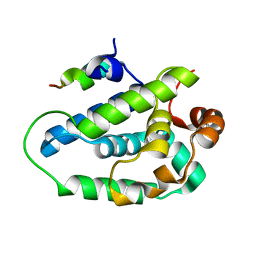 | | Peptide from EAEC T6SS Sci1 SciI protein | | Descriptor: | Peptide from EAEC T6SS Sci1 SciI protein, Putative type VI secretion protein | | Authors: | Douzi, B, Spinelli, S, Legrand, P, Lensi, V, Brunet, Y.R, Cascales, E, Cambillau, C. | | Deposit date: | 2012-10-09 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Role and specificity of ClpV ATPases in T6SS secretion.
To be Published
|
|
4HH5
 
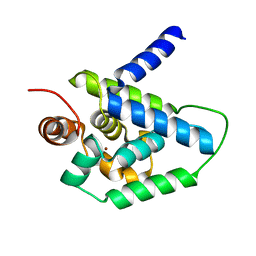 | | N-terminal domain (1-163) of ClpV1 ATPase from E.coli EAEC Sci1 T6SS. | | Descriptor: | BROMIDE ION, Putative type VI secretion protein | | Authors: | Douzi, B, Spinelli, S, Legrand, P, Lensi, V, Brunet, Y.R, Cascales, E, Cambillau, C. | | Deposit date: | 2012-10-09 | | Release date: | 2013-11-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role and specificity of ClpV ATPases in T6SS secretion.
To be Published
|
|
3FH2
 
 | |
5GUI
 
 | |
4Y0C
 
 | | The structure of Arabidopsis ClpT2 | | Descriptor: | CHLORIDE ION, Clp protease-related protein At4g12060, chloroplastic, ... | | Authors: | Kimber, M.S, Schultz, L. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structures, Functions, and Interactions of ClpT1 and ClpT2 in the Clp Protease System of Arabidopsis Chloroplasts.
Plant Cell, 27, 2015
|
|
5GKM
 
 | |
4Y0B
 
 | | The structure of Arabidopsis ClpT1 | | Descriptor: | CHLORIDE ION, Double Clp-N motif protein | | Authors: | Kimber, M.S, Schultz, L. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures, Functions, and Interactions of ClpT1 and ClpT2 in the Clp Protease System of Arabidopsis Chloroplasts.
Plant Cell, 27, 2015
|
|
5HBN
 
 | |
1R6C
 
 | | High resolution structure of ClpN | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpA | | Authors: | Xia, D, Maurizi, M.R, Guo, F, Singh, S.K, Esser, L. | | Deposit date: | 2003-10-15 | | Release date: | 2005-02-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic investigation of peptide binding sites in the N-domain of the
ClpA chaperone
J.Struct.Biol., 146, 2004
|
|
3WDB
 
 | |
3WDE
 
 | |
