4DOW
 
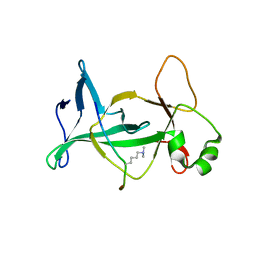 | | Structure of mouse ORC1 BAH domain bound to H4K20me2 | | Descriptor: | Histone H4, Origin recognition complex subunit 1 | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2012-02-10 | | Release date: | 2012-03-07 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome.
Nature, 484, 2012
|
|
8DS8
 
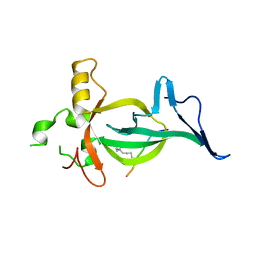 | |
1M4Z
 
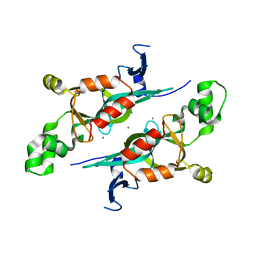 | | Crystal structure of the N-terminal BAH domain of Orc1p | | Descriptor: | MANGANESE (II) ION, ORIGIN RECOGNITION COMPLEX SUBUNIT 1 | | Authors: | Zhang, Z, Hayashi, M.K, Merkel, O, Stillman, B, Xu, R.-M. | | Deposit date: | 2002-07-05 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing.
EMBO J., 21, 2002
|
|
6OXB
 
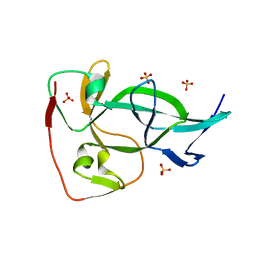 | |
2FVU
 
 | | Structure of the yeast Sir3 BAH domain | | Descriptor: | Regulatory protein SIR3 | | Authors: | Xu, R.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain.
Mol.Cell.Biol., 26, 2006
|
|
7LMK
 
 | |
7LMM
 
 | |
2FL7
 
 | | S. cerevisiae Sir3 BAH domain | | Descriptor: | Regulatory protein SIR3 | | Authors: | Keck, J.L, Hou, Z, Daner, J.R, Fox, C.A. | | Deposit date: | 2006-01-05 | | Release date: | 2006-05-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the Sir3 protein bromo adjacent homology (BAH) domain from S. cerevisiae at 1.95 A resolution.
Protein Sci., 15, 2006
|
|
4BB7
 
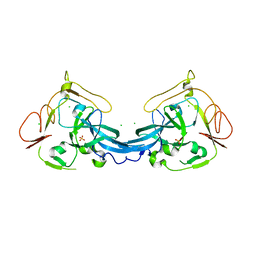 | | Crystal structure of the yeast Rsc2 BAH domain | | Descriptor: | CHLORIDE ION, CHROMATIN STRUCTURE-REMODELING COMPLEX SUBUNIT RSC2, GLYCEROL, ... | | Authors: | Chambers, A.L, Pearl, L.H, Oliver, A.W, Downs, J.A. | | Deposit date: | 2012-09-20 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Bah Domain of Rsc2 is a Histone H3 Binding Domain.
Nucleic Acids Res., 41, 2013
|
|
1W4S
 
 | |
4DOV
 
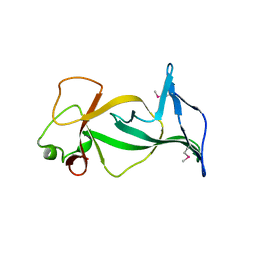 | | Structure of free mouse ORC1 BAH domain | | Descriptor: | Origin recognition complex subunit 1 | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2012-02-10 | | Release date: | 2012-03-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome.
Nature, 484, 2012
|
|
7XPJ
 
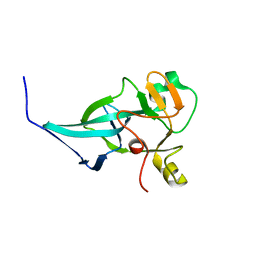 | | crystal structure of rice ASI1 BAH domain | | Descriptor: | BAH domain-containing protein | | Authors: | Yuan, J, Du, J. | | Deposit date: | 2022-05-04 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Molecular basis of locus-specific H3K9 methylation catalyzed by SUVH6 in plants.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7XPK
 
 | |
4KUL
 
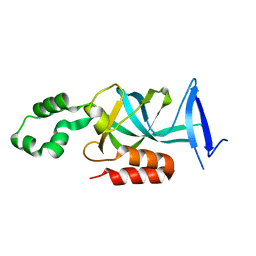 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain V83P mutant | | Descriptor: | Regulatory protein SIR3 | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4KUI
 
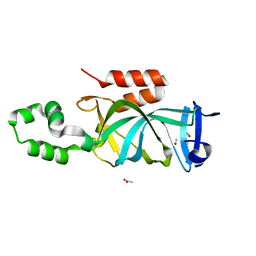 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain | | Descriptor: | ACETIC ACID, ISOPROPYL ALCOHOL, Regulatory protein SIR3 | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
7CCE
 
 | |
6VIL
 
 | |
8VLH
 
 | |
1ZHI
 
 | | Complex of the S. cerevisiae Orc1 and Sir1 interacting domains | | Descriptor: | Origin recognition complex subunit 1, Regulatory protein SIR1 | | Authors: | Hou, Z, Bernstein, D.A, Fox, C.A, Keck, J.L. | | Deposit date: | 2005-04-25 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the Sir1-origin recognition complex interaction in transcriptional silencing.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZBX
 
 | | Crystal structure of a Orc1p-Sir1p complex | | Descriptor: | Origin recognition complex subunit 1, Regulatory protein SIR1 | | Authors: | Hsu, H.C, Stillman, B, Xu, R.M. | | Deposit date: | 2005-04-09 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for origin recognition complex 1 protein-silence information regulator 1 protein interaction in epigenetic silencing
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
9MVY
 
 | | Crystal structure of ZMET2 in complex with unmethylated CTG DNA and a histone H3Kc9me2 peptide | | Descriptor: | C49 ssDNA, DNA (cytosine-5)-methyltransferase 1, Histone H3.2, ... | | Authors: | Herle, G, Fang, J, Song, J. | | Deposit date: | 2025-01-16 | | Release date: | 2025-05-21 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of an Unfavorable de Novo DNA Methylation Complex of Plant Methyltransferase ZMET2.
J.Mol.Biol., 437, 2025
|
|
7MCA
 
 | |
6V8O
 
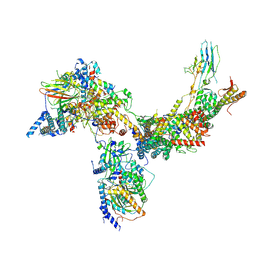 | | RSC core | | Descriptor: | Chromatin structure-remodeling complex protein RSC14, Chromatin structure-remodeling complex protein RSC3, Chromatin structure-remodeling complex protein RSC30, ... | | Authors: | Patel, A.B, Moore, C.M, Greber, B.J, Nogales, E. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement.
Elife, 8, 2019
|
|
4YOC
 
 | | Crystal Structure of human DNMT1 and USP7/HAUSP complex | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, Ubiquitin carboxyl-terminal hydrolase 7, ZINC ION | | Authors: | Cheng, J, Yang, H, Fang, J, Gong, R, Wang, P, Li, Z, Xu, Y. | | Deposit date: | 2015-03-11 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.916 Å) | | Cite: | Molecular mechanism for USP7-mediated DNMT1 stabilization by acetylation.
Nat Commun, 6, 2015
|
|
5GUT
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - N1248A | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Chen, S.J, Ye, F. | | Deposit date: | 2016-08-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
