3WDD
 
 | |
3WDC
 
 | |
3WDB
 
 | |
6CN8
 
 | | High-resolution structure of ClpC1-NTD binding to Rufomycin-I | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC1, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Abad-Zapatero, C, Wolf, N.W. | | Deposit date: | 2018-03-07 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Resolution Structure of ClpC1-Rufomycin and Ligand Binding Studies Provide a Framework to Design and Optimize Anti-Tuberculosis Leads.
Acs Infect Dis., 5, 2019
|
|
3WDE
 
 | |
8IBP
 
 | |
6PBQ
 
 | | Structure of ClpC1-NTD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ATP-dependent Clp protease ATP-binding subunit ClpC1, PHOSPHATE ION | | Authors: | Abad-Zapatero, C, Wolf, N.M. | | Deposit date: | 2019-06-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the N-terminal domain of ClpC1 in complex with the antituberculosis natural product ecumicin reveals unique binding interactions.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7AA4
 
 | | Structure of ClpC1-NTD bound to a CymA analogue | | Descriptor: | Negative regulator of genetic competence ClpC/mecB, polymer Cyclomarin A analogue | | Authors: | Meinhart, A, Morreale, F.E, Kaiser, M, Clausen, T. | | Deposit date: | 2020-09-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
6PBA
 
 | | Structure of ClpC1-NTD | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit ClpC1 | | Authors: | Abad-Zapatero, C, Wolf, N.M. | | Deposit date: | 2019-06-13 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of the N-terminal domain of ClpC1 in complex with the antituberculosis natural product ecumicin reveals unique binding interactions.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8IBO
 
 | |
8B9U
 
 | |
6UCR
 
 | | Structure of ClpC1-NTD L92S L96P | | Descriptor: | ACETATE ION, Negative regulator of genetic competence ClpC/mecB | | Authors: | Abad-Zapatero, C, Wolf, N.M. | | Deposit date: | 2019-09-17 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the N-terminal domain of ClpC1 in complex with the antituberculosis natural product ecumicin reveals unique binding interactions.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6PBS
 
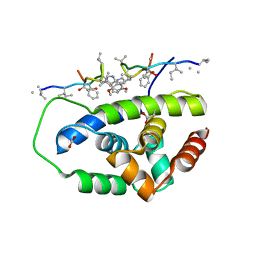 | | Structure of ClpC1-NTD in complex with Ecumicin | | Descriptor: | ACETATE ION, ATP-dependent Clp protease ATP-binding subunit ClpC1, ecumicin | | Authors: | Abad-Zapatero, C, Wolf, N.M. | | Deposit date: | 2019-06-14 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the N-terminal domain of ClpC1 in complex with the antituberculosis natural product ecumicin reveals unique binding interactions.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8A8V
 
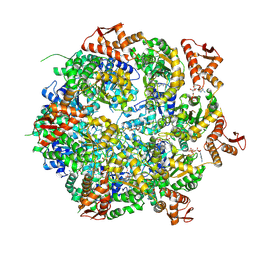 | | Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Cyclomarin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | Authors: | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
8A8U
 
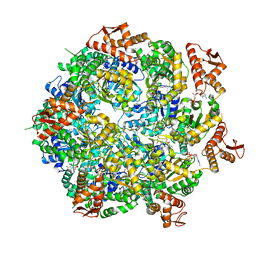 | | Mycobacterium tuberculosis ClpC1 hexamer structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | Authors: | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
8A8W
 
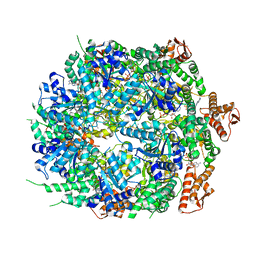 | | Mycobacterium tuberculosis ClpC1 hexamer structure bound to the natural product antibiotic Ecumycin (class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpC1, Bound polypeptide | | Authors: | Felix, J, Fraga, H, Gragera, M, Bueno, T, Weinhaeupl, K. | | Deposit date: | 2022-06-24 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Structure of the drug target ClpC1 unfoldase in action provides insights on antibiotic mechanism of action.
J.Biol.Chem., 298, 2022
|
|
