1K3A
 
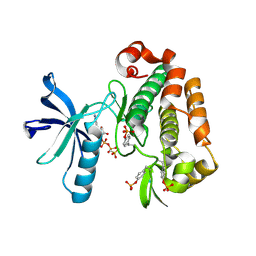 | | Structure of the Insulin-like Growth Factor 1 Receptor Kinase | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, insulin receptor substrate 1, insulin-like growth factor 1 receptor | | Authors: | Favelyukis, S, Till, J.H, Hubbard, S.R, Miller, W.T. | | Deposit date: | 2001-10-02 | | Release date: | 2001-11-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and autoregulation of the insulin-like growth factor 1 receptor kinase.
Nat.Struct.Biol., 8, 2001
|
|
5AXI
 
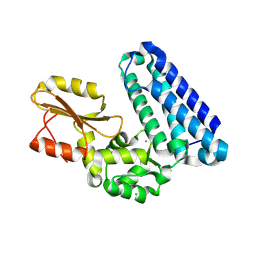 | | Crystal structure of Cbl-b TKB domain in complex with Cblin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cblin, ... | | Authors: | Ohno, A, Maita, N, Ochi, A, Nakao, R, Nikawa, T. | | Deposit date: | 2015-07-29 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the TKB domain of ubiquitin ligase Cbl-b complexed with its small inhibitory peptide, Cblin
Arch.Biochem.Biophys., 594, 2016
|
|
1QQG
 
 | |
1AYB
 
 | |
2Z8C
 
 | |
1IRS
 
 | | IRS-1 PTB DOMAIN COMPLEXED WITH A IL-4 RECEPTOR PHOSPHOPEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | IL-4 RECEPTOR PHOSPHOPEPTIDE, IRS-1 | | Authors: | Zhou, M.-M, Huang, B, Olejniczak, E.T, Meadows, R.P, Shuker, S.B, Miyazaki, M, Trub, T, Shoelson, S.E, Feisk, S.W. | | Deposit date: | 1996-03-22 | | Release date: | 1997-05-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis for IL-4 receptor phosphopeptide recognition by the IRS-1 PTB domain.
Nat.Struct.Biol., 3, 1996
|
|
6BNT
 
 | |
7PPL
 
 | | SHP2 catalytic domain in complex with IRS1 (625-639) phosphopeptide (pTyr-632, pSer-636) | | Descriptor: | ETHANOL, GLYCEROL, Insulin receptor substrate 1, ... | | Authors: | Sok, P, Zeke, A, Remenyi, A. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural insights into the pSer/pThr dependent regulation of the SHP2 tyrosine phosphatase in insulin and CD28 signaling.
Nat Commun, 13, 2022
|
|
7PPM
 
 | | SHP2 catalytic domain in complex with IRS1 (889-901) phosphopeptide (pSer-892, pTyr-896) | | Descriptor: | GLYCEROL, Insulin receptor substrate 1, Tyrosine-protein phosphatase non-receptor type 11,Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sok, P, Zeke, A, Remenyi, A. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural insights into the pSer/pThr dependent regulation of the SHP2 tyrosine phosphatase in insulin and CD28 signaling.
Nat Commun, 13, 2022
|
|
5U1M
 
 | |
