6MVX
 
 | |
7JHG
 
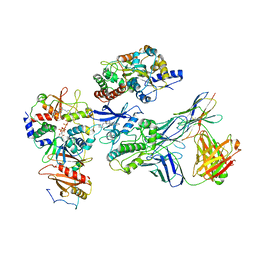 | | Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Dorsomorphin (Compound C) and Fab-nanobody | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Murkherjee, S, Zhou, X.E, Xu, T.H, Xu, H.E, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-21 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
8ZMR
 
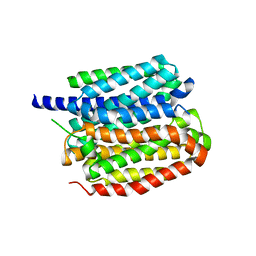 | | Vesamicol-bound VAChT | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Vesicular acetylcholine transporter,DARPinoff7, vesamicol | | Authors: | Zhang, Z, Zhang, Y, Dai, F, Zhang, Y.X, Lee, C.-H. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into VAChT neurotransmitter recognition and inhibition.
Cell Res., 2024
|
|
8HA1
 
 | |
7W9L
 
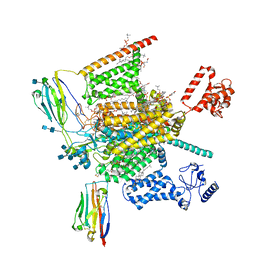 | | Cryo-EM structure of human Nav1.7(E406K)-beta1-beta2 complex | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | Deposit date: | 2021-12-10 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
4BLA
 
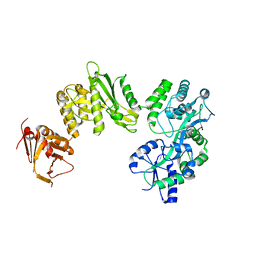 | | Crystal structure of full-length human Suppressor of fused (SUFU) mutant lacking a regulatory subdomain (crystal form II) | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG | | Authors: | Cherry, A.L, Finta, C, Karlstrom, M, Toftgard, R, Jovine, L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3IOT
 
 | |
3IOW
 
 | |
6VXO
 
 | | NaChBac-Nav1.7VSDII chimera in nanodisc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, NaChBac-Nav1.7VSDII chimera | | Authors: | Yan, N, Gao, S. | | Deposit date: | 2020-02-22 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Employing NaChBac for cryo-EM analysis of toxin action on voltage-gated Na+channels in nanodisc.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6HD9
 
 | | Crystal structure of the potassium channel MtTMEM175 with rubidium | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, RUBIDIUM ION, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
6NT4
 
 | | Cryo-EM structure of a human-cockroach hybrid Nav channel bound to alpha-scorpion toxin AaH2. | | Descriptor: | (7E,21R,24S)-27-amino-24-hydroxy-18,24-dioxo-19,23,25-trioxa-24lambda~5~-phosphaheptacos-7-en-21-yl (9Z,12E)-octadeca-9,12-dienoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Clairfeuille, T, Rohou, A, Payandeh, J. | | Deposit date: | 2019-01-28 | | Release date: | 2019-02-20 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of alpha-scorpion toxin action on Na v channels.
Science, 363, 2019
|
|
6X91
 
 | | Crystal structure of MBP-fused human APOBEC1 | | Descriptor: | CACODYLATE ION, Maltodextrin-binding protein, C->U-editing enzyme APOBEC-1 chimera, ... | | Authors: | Wolfe, A.D, Li, S.-X, Chen, X.S. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | The structure of APOBEC1 and insights into its RNA and DNA substrate selectivity.
NAR Cancer, 2, 2020
|
|
5EK0
 
 | | Human Nav1.7-VSD4-NavAb in complex with GX-936. | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-cyano-4-[2-[2-(1-ethylazetidin-3-yl)pyrazol-3-yl]-4-(trifluoromethyl)phenoxy]-~{N}-(1,2,4-thiadiazol-5-yl)benzenesulfonamide, Chimera of bacterial Ion transport protein and human Sodium channel protein type 9 subunit alpha | | Authors: | Ahuja, S, Mukund, S, Starovasnik, M.A, Koth, C.M, Payandeh, J. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist.
Science, 350, 2015
|
|
4XAJ
 
 | | Crystal structure of human NR2E1/TLX | | Descriptor: | Atrophin/grunge, Maltose-binding periplasmic protein,Nuclear receptor subfamily 2 group E member 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhi, X, Zhou, E, Xu, E. | | Deposit date: | 2014-12-14 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.551 Å) | | Cite: | Structural basis for corepressor assembly by the orphan nuclear receptor TLX.
Genes Dev., 29, 2015
|
|
4XZV
 
 | | Crystal Structure of SLMO1-TRIAP1 Complex | | Descriptor: | Maltose-binding periplasmic protein,TP53-regulated inhibitor of apoptosis 1, Protein slowmo homolog 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Miliara, X, Garnett, J.A, Matthews, S.J. | | Deposit date: | 2015-02-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Structural insight into the TRIAP1/PRELI-like domain family of mitochondrial phospholipid transfer complexes.
Embo Rep., 16, 2015
|
|
5GS2
 
 | | Crystal structure of diabody complex with repebody and MBP | | Descriptor: | Maltose-binding periplasmic protein, anti-MBP, anti-repebody, ... | | Authors: | Kim, J.H, Song, D.H, Youn, S.J, Kim, J.W, Cho, G, Lee, H, Lee, J.O. | | Deposit date: | 2016-08-13 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.592 Å) | | Cite: | Crystal structure of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Sci Rep, 6, 2016
|
|
3IOR
 
 | |
6N4Q
 
 | | CryoEM structure of Nav1.7 VSD2 (actived state) in complex with the gating modifier toxin ProTx2 | | Descriptor: | Beta/omega-theraphotoxin-Tp2a, Fab heavy chain, Fab light chain, ... | | Authors: | Xu, H, Rohou, A, Arthur, C.P, Estevez, A, Ciferri, C, Payandeh, J, Koth, C.M. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin.
Cell, 176, 2019
|
|
7K48
 
 | | Structure of NavAb/Nav1.7-VS2A chimera trapped in the resting state by tarantula toxin m3-Huwentoxin-IV | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Ion transport protein,Sodium channel protein type 9 subunit alpha chimera, Mu-theraphotoxin-Hs2a | | Authors: | Wisedchaisri, G, Tonggu, L, Gamal El-Din, T.M, McCord, E, Zheng, N, Catterall, W.A. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-02 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for High-Affinity Trapping of the Na V 1.7 Channel in Its Resting State by Tarantula Toxin.
Mol.Cell, 81, 2021
|
|
5W0Z
 
 | |
8HLB
 
 | | Cryo-EM structure of biparatopic antibody Bp109-92 in complex with TNFR2 | | Descriptor: | TR109 heavy chain, TR109 light chain, TR92 heavy chain, ... | | Authors: | Akiba, H, Fujita, J, Ise, T, Nishiyama, K, Miyata, T, Kato, T, Namba, K, Ohno, H, Kamada, H, Nagata, S, Tsumoto, K. | | Deposit date: | 2022-11-29 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Development of a 1:1-binding biparatopic anti-TNFR2 antagonist by reducing signaling activity through epitope selection.
Commun Biol, 6, 2023
|
|
8ZMS
 
 | | Acetylcholine-bound VAChT | | Descriptor: | ACETYLCHOLINE, Maltose/maltodextrin-binding periplasmic protein,Vesicular acetylcholine transporter,DARPinoff7 | | Authors: | Zhang, Z, Zhang, Y, Dai, F, Zhang, Y.X, Lee, C.-H. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into VAChT neurotransmitter recognition and inhibition.
Cell Res., 2024
|
|
3IOV
 
 | |
3IO6
 
 | | Huntingtin amino-terminal region with 17 Gln residues - crystal C92-a | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein, HUNTINGTIN FUSION PROTEIN, ... | | Authors: | Kim, M.W, Chelliah, Y, Kim, S.W, Otwinowski, Z, Bezprozvanny, I. | | Deposit date: | 2009-08-13 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Secondary structure of Huntingtin amino-terminal region.
Structure, 17, 2009
|
|
3IOU
 
 | |
