3F9X
 
 | | Structural Insights into Lysine Multiple Methylation by SET Domain Methyltransferases, SET8-Y334F / H4-Lys20me2 / AdoHcy | | Descriptor: | Histone H4, Histone-lysine N-methyltransferase SETD8, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Couture, J.-F, Dirk, L.M.A, Brunzelle, J.S, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural origins for the product specificity of SET domain protein methyltransferases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
8PEF
 
 | |
5TEG
 
 | |
3UVW
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a diacetylated histone 4 peptide (H4K5acK8ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Histone H4 | | Authors: | Filippakopoulos, P, Felletar, I, Picaud, S, Keates, T, Muniz, J, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-30 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
8UK5
 
 | | Crystal structure of the bromodomain of human ATAD2B in complex with histone H4S1(ph)K5ac | | Descriptor: | ATPase family AAA domain-containing protein 2B, Histone H4S1(ph)K5ac | | Authors: | Montgomery, C, Phillips, M, Nix, J.C, Glass, K.C. | | Deposit date: | 2023-10-12 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Impact of Combinatorial Histone Modifications on Acetyllysine Recognition by the ATAD2 and ATAD2B Bromodomains.
J.Med.Chem., 67, 2024
|
|
4YY6
 
 | | Crystal structure of BRD9 Bromodomain bound to a butyryllysine peptide | | Descriptor: | Bromodomain-containing protein 9, Histone H4 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
1ZKK
 
 | | Crystal structure of hSET8 in ternary complex with H4 peptide (16-24) and AdoHcy | | Descriptor: | Histone-lysine N-methyltransferase, H4 lysine-20 specific, Peptide corresponding to residues 15-24 of histone H4, ... | | Authors: | Couture, J.-F, Collazo, E, Brunzelle, J.S, Trievel, R.C. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase
Genes Dev., 19, 2005
|
|
5FFW
 
 | | Crystal structure of the bromodomain of human BRPF1 in complex with H4K5acK8ac histone peptide | | Descriptor: | Histone H4, Peregrin | | Authors: | Tallant, C, Nunez-Alonso, G, Savitsky, P, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S. | | Deposit date: | 2015-12-19 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the bromodomain of human BRPF1 in complex with H4K5acK8ac histone peptide
To Be Published
|
|
2BQZ
 
 | | Crystal structure of a ternary complex of the human histone methyltransferase Pr-SET7 (also known as SET8) | | Descriptor: | HISTONE H4, S-ADENOSYL-L-HOMOCYSTEINE, SET8 PROTEIN | | Authors: | Xiao, B, Jing, C, Kelly, G, Walker, P.A, Muskett, F.W, Frenkiel, T.A, Martin, S.R, Sarma, K, Reinberg, D, Gamblin, S.J, Wilson, J.R. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Specificity and Mechanism of the Histone Methyltransferase Pr-Set7
Genes Dev., 19, 2005
|
|
3QZT
 
 | | Crystal Structure of BPTF bromo in complex with histone H4K16ac - Form II | | Descriptor: | GLYCEROL, Histone H4, Nucleosome-remodeling factor subunit BPTF | | Authors: | Li, H, Ruthenburg, A.J, Patel, D.J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions.
Cell(Cambridge,Mass.), 145, 2011
|
|
4YYI
 
 | | Crystal structure of BRD9 Bromodomain bound to an acetylated peptide | | Descriptor: | Bromodomain-containing protein 9, Histone H4 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
4YYM
 
 | | Crystal structure of TAF1 BD2 Bromodomain bound to a butyryllysine peptide | | Descriptor: | CALCIUM ION, Histone H4, Transcription initiation factor TFIID subunit 1 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
3F9Y
 
 | | Structural Insights into Lysine Multiple Methylation by SET Domain Methyltransferases, SET8-Y334F / H4-Lys20me1 / AdoHcy | | Descriptor: | Histone H4, Histone-lysine N-methyltransferase SETD8, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Couture, J.-F, Dirk, L.M.A, Brunzelle, J.S, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural origins for the product specificity of SET domain protein methyltransferases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4YYD
 
 | | Crystal structure of BRD9 Bromodomain bound to a crotonyllysine peptide | | Descriptor: | Bromodomain-containing protein 9, Histone H4 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
8B5C
 
 | | Human BRD4 bromdomain 1 in complex with a H4 peptide containing ApmTri (H4K5/8ApmTri) | | Descriptor: | Bromodomain-containing protein 4, H4K5/8ApmTri | | Authors: | Braun, M.B, Bartlick, N, Stehle, T. | | Deposit date: | 2022-09-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Synthesis, Biochemical Characterization, and Genetic Encoding of a 1,2,4-Triazole Amino Acid as an Acetyllysine Mimic for Bromodomains of the BET Family.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6RXS
 
 | | Crystal structure of CobB Ac3(A76G,Y92A, I131L, V187Y) in complex with H4K16-Acetyl peptide | | Descriptor: | GLYCEROL, Histone H4, NAD-dependent protein deacylase, ... | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7M98
 
 | | ATAD2 bromodomain complexed with histone H4K5ac (res 1-10) ligand | | Descriptor: | ATPase family AAA domain-containing protein 2, Histone H4 | | Authors: | Malone, K.L, Phillips, M, Nix, J.C, Glass, K.C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Coordination of Di-Acetylated Histone Ligands by the ATAD2 Bromodomain.
Int J Mol Sci, 22, 2021
|
|
6VO5
 
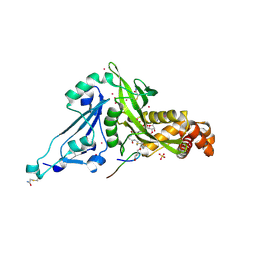 | | Crystal structure of Human histone acetytransferas 1 (HAT1) in complex with isobutryl-COA and K12A mutant variant of histone H4 | | Descriptor: | ACETATE ION, GLYCEROL, Histone H4, ... | | Authors: | Halabelian, L, Zeng, H, Dong, A, Loppnau, P, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Human histone acetytransferas 1 (HAT1) in complex with isobutryl-COA and K12A mutant variant of histone H4
to be published
|
|
3F9W
 
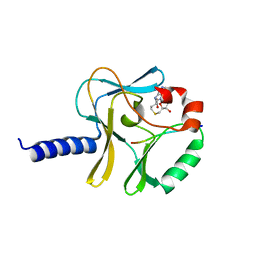 | | Structural Insights into Lysine Multiple Methylation by SET Domain Methyltransferases, SET8-Y334F / H4-Lys20 / AdoHcy | | Descriptor: | Histone H4, Histone-lysine N-methyltransferase SETD8, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Couture, J.-F, Dirk, L.M.A, Brunzelle, J.S, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural origins for the product specificity of SET domain protein methyltransferases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3F9Z
 
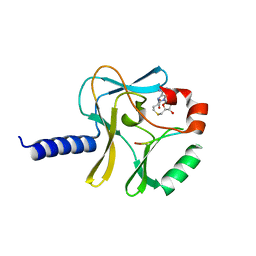 | | Structural Insights into Lysine Multiple Methylation by SET Domain Methyltransferases, SET8-Y245F / H4-Lys20 / AdoHcy | | Descriptor: | Histone H4, Histone-lysine N-methyltransferase SETD8, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Couture, J.-F, Dirk, L.M.A, Brunzelle, J.S, Houtz, R.L, Trievel, R.C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural origins for the product specificity of SET domain protein methyltransferases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5YE3
 
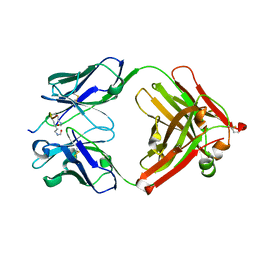 | | Crystal structure of the complex of di-acetylated histone H4 and 2A7D9 Fab fragment | | Descriptor: | 2A7D9 L chain, 2A7D9 VH CH1 chain, di-acetylated histone H4 | | Authors: | Matsuda, T, Ito, T, Wakamori, M, Umehara, T. | | Deposit date: | 2017-09-15 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states.
Epigenetics, 13, 2018
|
|
2IG0
 
 | | Structure of 53BP1/methylated histone peptide complex | | Descriptor: | Dimethylated Histone H4-K20 peptide, SULFATE ION, Tumor suppressor p53-binding protein 1 | | Authors: | Mer, G. | | Deposit date: | 2006-09-22 | | Release date: | 2007-01-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Methylation State-Specific Recognition of Histone H4-K20 by 53BP1 and Crb2 in DNA Repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
3O36
 
 | |
4QUT
 
 | | Structure of the bromodomain of human ATPase family AAA domain-containing protein 2 (ATAD2) complexed with Histone H4-K(ac)12 | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, Histone H4, ... | | Authors: | Chaikuad, A, Felletar, I, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-12 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Atad2 is a generalist facilitator of chromatin dynamics in embryonic stem cells.
J Mol Cell Biol, 8, 2016
|
|
4YYH
 
 | | Crystal structure of BRD9 Bromodomain bound to a crotonyllysine peptide | | Descriptor: | Bromodomain-containing protein 9, Histone H4 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
