4D0M
 
 | | Phosphatidylinositol 4-kinase III beta in a complex with Rab11a-GTP- gamma-S and the Rab-binding domain of FIP3 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, N-(5-(4-CHLORO-3-(2-HYDROXY-ETHYLSULFAMOYL)- PHENYLTHIAZOLE-2-YL)-ACETAMIDE, ... | | Authors: | Burke, J.E, Inglis, A.J, Perisic, O, Masson, G.R, McLaughlin, S.H, Rutaganira, F, Shokat, K.M, Williams, R.L. | | Deposit date: | 2014-04-29 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structures of Pi4Kiiibeta Complexes Show Simultaneous Recruitment of Rab11 and its Effectors.
Science, 344, 2014
|
|
1IF0
 
 | | PSEUDO-ATOMIC MODEL OF BACTERIOPHAGE HK97 PROCAPSID (PROHEAD II) | | Descriptor: | PROTEIN (MAJOR CAPSID PROTEIN GP5) | | Authors: | Conway, J.F, Wikoff, W.R, Cheng, N, Duda, R.L, Hendrix, R.W, Johnson, J.E, Steven, A.C. | | Deposit date: | 2001-04-11 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Virus maturation involving large subunit rotations and local refolding.
Science, 292, 2001
|
|
1I50
 
 | | RNA POLYMERASE II CRYSTAL FORM II AT 2.8 A RESOLUTION | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.5KD POLYPEPTIDE, ... | | Authors: | Cramer, P, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2001-02-23 | | Release date: | 2001-04-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution.
Science, 292, 2001
|
|
1I7E
 
 | | C-Terminal Domain Of Mouse Brain Tubby Protein bound to Phosphatidylinositol 4,5-bis-phosphate | | Descriptor: | L-ALPHA-GLYCEROPHOSPHO-D-MYO-INOSITOL-4,5-BIS-PHOSPHATE, TUBBY PROTEIN | | Authors: | Santagata, S, Boggon, T.J, Baird, C.L, Shan, W.S, Shapiro, L. | | Deposit date: | 2001-03-08 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | G-protein signaling through tubby proteins.
Science, 292, 2001
|
|
1LDF
 
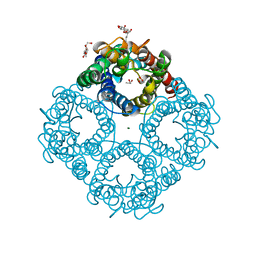 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) MUTATION W48F, F200T | | Descriptor: | GLYCEROL, Glycerol uptake facilitator protein, MAGNESIUM ION, ... | | Authors: | Nollert, P, Miercke, L.J.W, O'Connell, J, Stroud, R.M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Control of the selectivity of the aquaporin water channel family by global orientational tuning.
Science, 296, 2002
|
|
1LDA
 
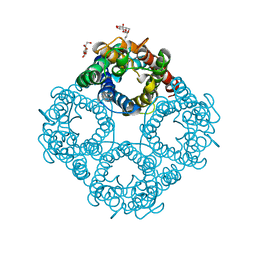 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) WITHOUT SUBSTRATE GLYCEROL | | Descriptor: | Glycerol uptake facilitator protein, octyl beta-D-glucopyranoside | | Authors: | Nollert, P, Miercke, L.J.W, O'Connell, J, Stroud, R.M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Control of the selectivity of the aquaporin water channel family by global orientational tuning.
Science, 296, 2002
|
|
1LDI
 
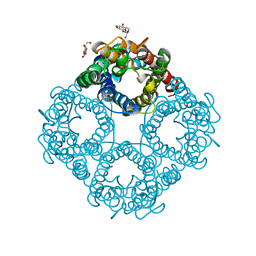 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) WITHOUT SUBSTRATE GLYCEROL | | Descriptor: | Glycerol uptake facilitator protein, octyl beta-D-glucopyranoside | | Authors: | Nollert, P, Miercke, L.J.W, O'Connell, J, Stroud, R.M. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Control of the selectivity of the aquaporin water channel family by global orientational tuning.
Science, 296, 2002
|
|
5V8K
 
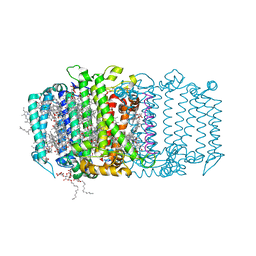 | | Homodimeric reaction center of H. modesticaldum | | Descriptor: | 1-[GLYCEROLYLPHOSPHONYL]-2-[8-(2-HEXYL-CYCLOPROPYL)-OCTANAL-1-YL]-3-[HEXADECANAL-1-YL]-GLYCEROL, 4,4'-Diaponeurosporene, 8(1)-OH-Chlorophyll aF, ... | | Authors: | Gisriel, C, Sarrou, I, Ferlez, B, Golbeck, J, Redding, K.E, Fromme, R. | | Deposit date: | 2017-03-22 | | Release date: | 2017-09-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a symmetric photosynthetic reaction center-photosystem.
Science, 357, 2017
|
|
7O7Z
 
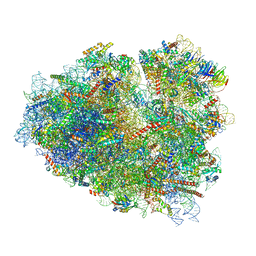 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (classified for pseudoknot) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7NWL
 
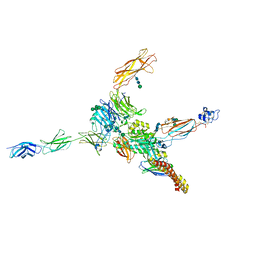 | | Cryo-EM structure of human integrin alpha5beta1 (open form) in complex with fibronectin and TS2/16 Fv-clasp | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-5, ... | | Authors: | Schumacher, S, Dedden, D, Vazquez Nunez, R, Matoba, K, Takagi, J, Biertumpfel, C, Mizuno, N. | | Deposit date: | 2021-03-17 | | Release date: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into integrin alpha 5 beta 1 opening by fibronectin ligand.
Sci Adv, 7, 2021
|
|
7O81
 
 | | Rabbit 80S ribosome colliding in another ribosome stalled by the SARS-CoV-2 pseudoknot | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O7Y
 
 | | Rabbit 80S ribosome stalled close to the mutated SARS-CoV-2 slippery site by a pseudoknot (high resolution) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
7O80
 
 | | Rabbit 80S ribosome in complex with eRF1 and ABCE1 stalled at the STOP codon in the mutated SARS-CoV-2 slippery site | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Bhatt, P.R, Scaiola, A, Leibundgut, M.A, Atkins, J.F, Ban, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-06-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of ribosomal frameshifting during translation of the SARS-CoV-2 RNA genome.
Science, 372, 2021
|
|
2KFU
 
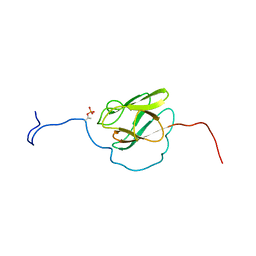 | | PknB-phosphorylated Rv1827 | | Descriptor: | Rv1827 pThr 22 | | Authors: | Smerdon, S.J, Nott, T.J, Kelly, G. | | Deposit date: | 2009-02-27 | | Release date: | 2009-06-16 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | An intramolecular switch regulates phosphoindependent FHA domain interactions in Mycobacterium tuberculosis.
Sci.Signal., 2, 2009
|
|
7O7F
 
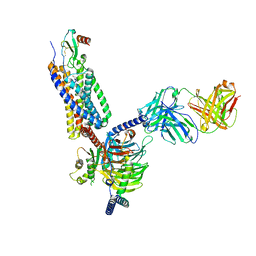 | | Structural basis of the activation of the CC chemokine receptor 5 by a chemokine agonist | | Descriptor: | C-C chemokine receptor type 5, C-C motif chemokine 5, Fab antibody fragment heavy chain, ... | | Authors: | Isaikina, P, Tsai, C.-J, Dietz, N.B, Pamula, F, Goldie, K.N, Schertler, G.F.X, Maier, T, Stahlberg, H, Deupi, X, Grzesiek, S. | | Deposit date: | 2021-04-13 | | Release date: | 2021-06-30 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of the activation of the CC chemokine receptor 5 by a chemokine agonist.
Sci Adv, 7, 2021
|
|
2KVQ
 
 | |
2KZG
 
 | | A Transient and Low Populated Protein Folding Intermediate at Atomic Resolution | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Korzhnev, D.M, Religa, T.L, Banachewicz, W, Fersht, A.R, Kay, L.E. | | Deposit date: | 2010-06-17 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A transient and low-populated protein-folding intermediate at atomic resolution.
Science, 329, 2010
|
|
2KS6
 
 | |
2L0J
 
 | | Solid State NMR structure of the M2 proton channel from Influenza A Virus in hydrated lipid bilayer | | Descriptor: | Matrix protein 2 | | Authors: | Sharma, M, Yi, M, Dong, H, Qin, H, Peterson, E, Busath, D.D, Zhou, H.X, Cross, T.A. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Insight into the mechanism of the influenza a proton channel from a structure in a lipid bilayer.
Science, 330, 2010
|
|
2L6S
 
 | | Efficacy of an HIV-1 entry inhibitor targeting the GP41 fusion peptide | | Descriptor: | VIR-576 | | Authors: | Forssmann, W, The, Y, Stoll, M, Adermann, K, Albrecht, U, Barlos, K, Busmann, A, Canales-Mayordomo, A, Gimenez-Gallego, G, Hirsch, J, Jimenez-Barbero, J, Meyer-Olson, D, Muench, J, Perez-Castells, J, Standker, L, Kirchhoff, F, Schmidt, R.E. | | Deposit date: | 2010-11-24 | | Release date: | 2011-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Short-term monotherapy in HIV-infected patients with a virus entry inhibitor against the gp41 fusion peptide.
Sci Transl Med, 2, 2010
|
|
9B6H
 
 | | Cryo-EM structure of the mouse TRPM8 channel in complex with the antagonist TC-I 2014 and the cooling agonist C3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-{7-(trifluoromethyl)-5-[2-(trifluoromethyl)phenyl]-1H-benzimidazol-2-yl}-1-oxa-2-azaspiro[4.5]dec-2-ene, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Yin, Y, Park, C.-G, Zhang, F, Fedor, J, Feng, S, Suo, Y, Im, W, Lee, S.-Y. | | Deposit date: | 2024-03-25 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Mechanisms of sensory adaptation and inhibition of the cold and menthol receptor TRPM8.
Sci Adv, 10, 2024
|
|
9C0J
 
 | |
9C8U
 
 | | Human PRC2 - RvLEAM (short) (1:6 molar ratio), cross-linked 10 min | | Descriptor: | Isoform 2 of Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, Polycomb protein SUZ12, ... | | Authors: | Abe, K.M, Li, G, He, Q, Grant, T, Lim, C. | | Deposit date: | 2024-06-13 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Human PRC2 - RvLEAM (short) (1:6 molar ratio), cross-linked 10 min
To Be Published
|
|
9EZY
 
 | | Vibrio cholerae DdmD-DdmE holo complex | | Descriptor: | 14 nucleotide DNA guide with terminal 5' phosphate, Helicase/UvrB N-terminal domain-containing protein, MAGNESIUM ION, ... | | Authors: | Loeff, L, Jinek, M. | | Deposit date: | 2024-04-14 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Molecular mechanism of plasmid elimination by the DdmDE defense system.
Science, 385, 2024
|
|
4UOS
 
 | | Thermodynamic hyperstability in parametrically designed helical bundles | | Descriptor: | DESIGNED HELICAL BUNDLE | | Authors: | Oberdorfer, G, Huang, P, Pei, X.Y, Xu, C, Gonen, T, Nannenga, B, DiMaio, D, Rogers, J, Luisi, B.F, Baker, D. | | Deposit date: | 2014-06-09 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | High Thermodynamic Stability of Parametrically Designed Helical Bundles
Science, 346, 2014
|
|
