1LPG
 
 | | CRYSTAL STRUCTURE OF FXA IN COMPLEX WITH 79. | | Descriptor: | Blood coagulation factor Xa, CALCIUM ION, [4-({[5-BENZYLOXY-1-(3-CARBAMIMIDOYL-BENZYL)-1H-INDOLE-2-CARBONYL]-AMINO}-METHYL)-PHENYL]-TRIMETHYL-AMMONIUM | | Authors: | Schreuder, H.A, Brachvogel, V, Liesum, A. | | Deposit date: | 2002-05-08 | | Release date: | 2003-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Quantitative Structure-Activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa.
J.Med.Chem., 45, 2002
|
|
1LPH
 
 | | LYS(B28)PRO(B29)-HUMAN INSULIN | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Ciszak, E, Beals, J.M, Frank, B.H, Baker, J.C, Carter, N.D, Smith, G.D. | | Deposit date: | 1995-04-19 | | Release date: | 1996-06-20 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of C-terminal B-chain residues in insulin assembly: the structure of hexameric LysB28ProB29-human insulin.
Structure, 3, 1995
|
|
1LPI
 
 | | HEW LYSOZYME: TRP...NA CATION-PI INTERACTION | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Wouters, J. | | Deposit date: | 1998-04-15 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cation-pi (Na+-Trp) interactions in the crystal structure of tetragonal lysozyme.
Protein Sci., 7, 1998
|
|
1LPJ
 
 | | Human cRBP IV | | Descriptor: | Retinol-binding protein IV, cellular | | Authors: | Calderone, V, Zanotti, G, Berni, R, Folli, C. | | Deposit date: | 2002-05-08 | | Release date: | 2003-01-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand binding and structural analysis of a human putative cellular retinol-binding protein
J.Biol.Chem., 277, 2002
|
|
1LPK
 
 | | CRYSTAL STRUCTURE OF FXA IN COMPLEX WITH 125. | | Descriptor: | 1-(3-CARBAMIMIDOYL-BENZYL)-1H-INDOLE-2-CARBOXYLIC ACID 3-CARBAMIMIDOYL-BENZYLESTER, Blood coagulation factor Xa, CALCIUM ION | | Authors: | Schreuder, H.A, Loenze, P, Brachvogel, V, Liesum, A. | | Deposit date: | 2002-05-08 | | Release date: | 2003-05-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and Quantitative Structure-Activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa
J.Med.Chem., 45, 2002
|
|
1LPL
 
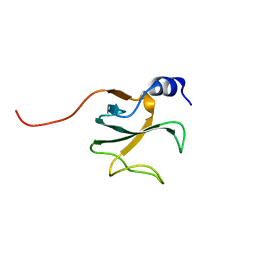 | | Structural Genomics of Caenorhabditis elegans: CAP-Gly domain of F53F4.3 | | Descriptor: | Hypothetical 25.4 kDa protein F53F4.3 in chromosome V | | Authors: | Li, S, Finley, J, Liu, Z.-J, Qiu, S.H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, DeLucas, L.J, Wang, B.-C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2002-05-08 | | Release date: | 2002-05-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of the
Cytoskeleton-associated Protein
Glycine-rich (CAP-Gly) Domain
J.Biol.Chem., 277, 2002
|
|
1LPM
 
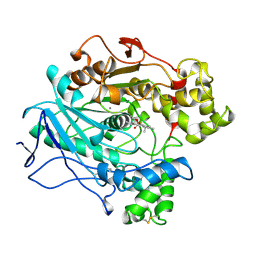 | | A STRUCTURAL BASIS FOR THE CHIRAL PREFERENCES OF LIPASES | | Descriptor: | (1R)-MENTHYL HEXYL PHOSPHONATE GROUP, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grochulski, P.G, Cygler, M.C. | | Deposit date: | 1995-01-06 | | Release date: | 1995-04-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A Structural Basis for the Chiral Preferences of Lipases
J.Am.Chem.Soc., 116, 1994
|
|
1LPN
 
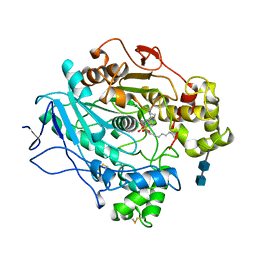 | |
1LPO
 
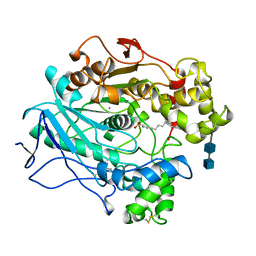 | |
1LPP
 
 | |
1LPQ
 
 | | Human DNA Topoisomerase I (70 Kda) In Non-Covalent Complex With A 22 Base Pair DNA Duplex Containing an 8-oxoG Lesion | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*(8OG)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', 5'-D(*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*CP*AP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3', DNA topoisomerase I | | Authors: | Lesher, D.T, Pommier, Y, Stewart, L, Redinbo, M.R. | | Deposit date: | 2002-05-08 | | Release date: | 2002-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | 8-Oxoguanine rearranges the active site of human topoisomerase I
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1LPS
 
 | | A STRUCTURAL BASIS FOR THE CHIRAL PREFERENCES OF LIPASES | | Descriptor: | (1S)-MENTHYL HEXYL PHOSPHONATE GROUP, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Grochulski, P.G, Cygler, M.C. | | Deposit date: | 1995-01-05 | | Release date: | 1995-02-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A Structural Basis for the Chiral Preferences of Lipases
J.Am.Chem.Soc., 116, 1994
|
|
1LPU
 
 | | Low Temperature Crystal Structure of the Apo-form of the catalytic subunit of protein kinase CK2 from Zea mays | | Descriptor: | BENZAMIDINE, Protein kinase CK2 | | Authors: | Niefind, K, Puetter, M, Guerra, B, Issinger, O.-G, Schomburg, D. | | Deposit date: | 2002-05-08 | | Release date: | 2002-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Inclining the purine base binding plane in protein kinase CK2 by exchanging the flanking side-chains generates a preference for ATP as a cosubstrate.
J.Mol.Biol., 347, 2005
|
|
1LPV
 
 | | DROSOPHILA MELANOGASTER DOUBLESEX (DSX), NMR, 18 STRUCTURES | | Descriptor: | Doublesex protein, ZINC ION | | Authors: | Zhu, L, Wilken, J, Phillips, N, Narendra, U, Chan, G, Stratton, S, Kent, S, Weiss, M.A. | | Deposit date: | 2002-05-08 | | Release date: | 2002-10-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers.
Genes Dev., 14, 2000
|
|
1LPW
 
 | |
1LPY
 
 | | Multiple Methionine Substitutions in T4 Lysozyme | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME, ... | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H.M, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-05-08 | | Release date: | 2002-05-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability.
Biophys.Chem., 100, 2003
|
|
1LPZ
 
 | | CRYSTAL STRUCTURE OF FXA IN COMPLEX WITH 41. | | Descriptor: | 1-(3-carbamimidoylbenzyl)-N-(3,5-dichlorobenzyl)-4-methyl-1H-indole-2-carboxamide, Blood coagulation factor Xa, CALCIUM ION | | Authors: | Schreuder, H.A, Brachvogel, V, Liesum, A. | | Deposit date: | 2002-05-08 | | Release date: | 2003-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Quantitative Structure-Activity relationship of 3-amidinobenzyl-1H-indole-2-carboxamides as potent, nonchiral, and selective inhibitors of blood coagulation factor Xa
J.Biol.Chem., 45, 2002
|
|
1LQ0
 
 | |
1LQ1
 
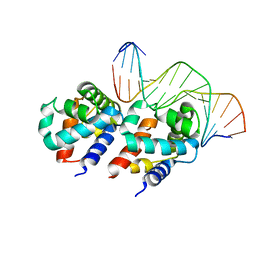 | | DNA Complexed Structure of the Key Transcription Factor Initiating Development in Sporulation Bacteria | | Descriptor: | 5'-D(*AP*CP*AP*AP*AP*AP*TP*TP*CP*GP*AP*CP*AP*CP*GP*A)-3', 5'-D(*TP*TP*CP*GP*TP*GP*TP*CP*GP*AP*AP*TP*TP*TP*TP*G)-3', Stage 0 sporulation protein A | | Authors: | Zhao, H, Msadek, T, Zapf, J, Madhusudan, Hoch, J.A, Varughese, K.I. | | Deposit date: | 2002-05-08 | | Release date: | 2002-08-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA complexed structure of the key transcription factor initiating development in sporulating bacteria.
Structure, 10, 2002
|
|
1LQ2
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with gluco-phenylimidazole | | Descriptor: | (5R,6R,7S,8S)-5-(HYDROXYMETHYL)-2-PHENYL-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hrmova, M, De Gori, R, Smith, B.J, Vasella, A, Varghese, J.N, Fincher, G.B. | | Deposit date: | 2002-05-09 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional Structure of the Barley {beta}-D-Glucan Glucohydrolase in Complex with a Transition State Mimic.
J.Biol.Chem., 279, 2004
|
|
1LQ7
 
 | | De Novo Designed Protein Model of Radical Enzymes | | Descriptor: | Alpha3W | | Authors: | Dai, Q.-H, Tommos, C, Fuentes, E.J, Blomberg, M, Dutton, P.L, Wand, A.J. | | Deposit date: | 2002-05-09 | | Release date: | 2002-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a De Novo Designed Protein Model of Radical Enzymes
J.Am.Chem.Soc., 124, 2002
|
|
1LQ8
 
 | | Crystal structure of cleaved protein C inhibitor | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Huntington, J.A, Kjellberg, M, Stenflo, J. | | Deposit date: | 2002-05-09 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Protein C Inhibitor Provides Insights into Hormone Binding and Heparin Activation
Structure, 11, 2003
|
|
1LQ9
 
 | | Crystal Structure of a Monooxygenase from the Gene ActVA-Orf6 of Streptomyces coelicolor Strain A3(2) | | Descriptor: | ACTVA-ORF6 MONOOXYGENASE, TETRAETHYLENE GLYCOL | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-05-09 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis
EMBO J., 22, 2003
|
|
1LQA
 
 | | TAS PROTEIN FROM ESCHERICHIA COLI IN COMPLEX WITH NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Tas protein | | Authors: | Obmolova, G, Teplyakov, A, Khil, P.P, Howard, A.J, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2002-05-09 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Crystal structure of the Escherichia coli Tas protein, an NADP(H)-dependent aldo-keto reductase
PROTEINS: STRUCT.,FUNCT.,GENET., 53, 2003
|
|
1LQB
 
 | | Crystal structure of a hydroxylated HIF-1 alpha peptide bound to the pVHL/elongin-C/elongin-B complex | | Descriptor: | Elongin B, Elongin C, Hypoxia-inducible factor 1 ALPHA, ... | | Authors: | Hon, W.C, Wilson, M.I, Harlos, K, Claridge, T.D, Schofield, C.J, Pugh, C.W, Maxwell, P.H, Ratcliffe, P.J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2002-05-09 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL.
Nature, 417, 2002
|
|
