5KNI
 
 | |
5N2O
 
 | | Structure Of P63 SAM Domain L514F Mutant Causative Of AEC Syndrome | | Descriptor: | Tumor protein 63 | | Authors: | Rinnenthal, J, Wuerz, J.M, Osterburg, C, Guentert, P, Doetsch, V. | | Deposit date: | 2017-02-08 | | Release date: | 2018-02-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein aggregation of the p63 transcription factor underlies severe skin fragility in AEC syndrome.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1V38
 
 | | Solution structure of the Sterile Alpha Motif (SAM) domain of mouse SAMSN1 | | Descriptor: | SAM-domain protein SAMSN-1 | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-29 | | Release date: | 2004-04-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Sterile Alpha Motif (SAM) domain of mouse SAMSN1
To be Published
|
|
7Z72
 
 | | Crystal structure of p63 SAM in complex with darpin A5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Darpin A5, Isoform 9 of Tumor protein 63 | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
6QWV
 
 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Isupov, N.M, Opatowsky, Y. | | Deposit date: | 2019-03-06 | | Release date: | 2019-07-03 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural Evidence for an Octameric Ring Arrangement of SARM1.
J.Mol.Biol., 431, 2019
|
|
1B0X
 
 | |
3H8M
 
 | | SAM domain of human ephrin type-a receptor 7 (EPHA7) | | Descriptor: | Ephrin type-A receptor 7 | | Authors: | Walker, J.R, Yermekbayeva, L, Butler-Cole, C, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-29 | | Release date: | 2009-05-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SAM Domain of Human Ephrin Type-A Receptor 7 (Epha7)
To be Published
|
|
1KW4
 
 | |
1COK
 
 | |
1DXS
 
 | |
1X9X
 
 | | Solution Structure of Dimeric SAM Domain from MAPKKK Ste11 | | Descriptor: | Serine/threonine-protein kinase STE11 | | Authors: | Bhattacharjya, S, Xu, P, Gingras, R, Shaykhutdinov, R, Wu, C, Whiteway, M, Ni, F. | | Deposit date: | 2004-08-24 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the dimeric SAM domain of MAPKKK Ste11 and its interactions with the adaptor protein Ste50 from the budding yeast: implications for Ste11 activation and signal transmission through the Ste50-Ste11 complex.
J.Mol.Biol., 344, 2004
|
|
2B6G
 
 | |
1OW5
 
 | |
6O0S
 
 | | Crystal structure of the tandem SAM domains from human SARM1 | | Descriptor: | Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6O0T
 
 | | Crystal structure of selenomethionine labelled tandem SAM domains (L446M:L505M:L523M mutant) from human SARM1 | | Descriptor: | Sterile alpha and TIR motif-containing protein 1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Deerain, N, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
6PW7
 
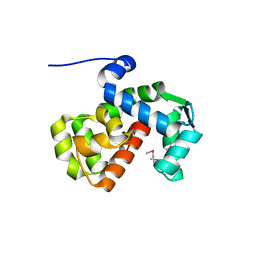 | | X-ray crystal structure of C. elegans STIM EF-SAM domain | | Descriptor: | CALCIUM ION, Stromal interaction molecule 1 | | Authors: | Enomoto, M, Nishikawa, T, Back, S.I, Ishiyama, N, Zheng, L, Stathopulos, P.B, Ikura, M. | | Deposit date: | 2019-07-22 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Coordination of a Single Calcium Ion in the EF-hand Maintains the Off State of the Stromal Interaction Molecule Luminal Domain.
J.Mol.Biol., 432, 2020
|
|
5JU5
 
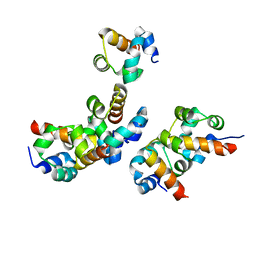 | |
5JTI
 
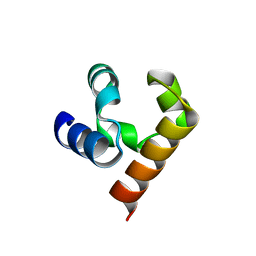 | |
5JRT
 
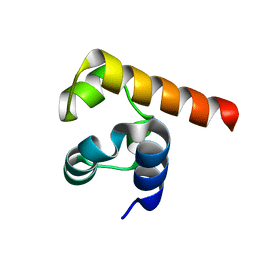 | |
1RG6
 
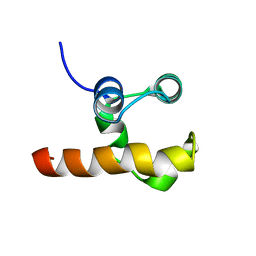 | | Solution structure of the C-terminal domain of p63 | | Descriptor: | second splice variant p63 | | Authors: | Cadot, B, Candi, E, Cicero, D.O, Desideri, A, Mele, S, Melino, G, Paci, M. | | Deposit date: | 2003-11-11 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of p63
To be Published
|
|
6ZG1
 
 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
2Y9T
 
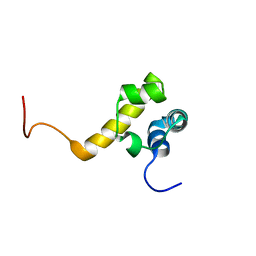 | |
3BS7
 
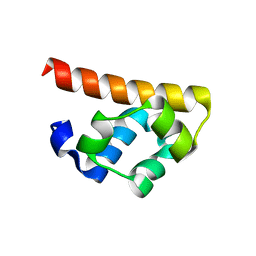 | |
2Y9U
 
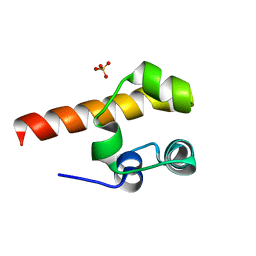 | | Structural basis of p63a SAM domain mutants involved in AEC syndrome | | Descriptor: | SULFATE ION, TUMOR PROTEIN 63 | | Authors: | Sathyamurthy, A, Freund, S.M.V, Johnson, C.M, Allen, M.D. | | Deposit date: | 2011-02-16 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of P63Alpha Sam Domain Mutants Involved in Aec Syndrome.
FEBS J., 278, 2011
|
|
3BQ7
 
 | |
