7Q8E
 
 | |
7QT4
 
 | | Antibody FenAb709 - fentanyl complex | | Descriptor: | 2-[2-[2-[2-[[5-oxidanylidene-5-[2-[4-[phenyl(propanoyl)amino]piperidin-1-yl]ethylamino]pentanoyl]amino]ethanoylamino]ethanoylamino]ethanoylamino]ethanoic acid, Antibody heavy chain, Antibody light chain | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E. | | Deposit date: | 2022-01-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A trypanosome-derived immunotherapeutics platform elicits potent high-affinity antibodies, negating the effects of the synthetic opioid fentanyl.
Cell Rep, 42, 2023
|
|
7ZU3
 
 | |
7M0Y
 
 | | Crystal structure of the BRAF:MEK1 kinases in complex with AMPPNP and Trametinib | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, K, Gonzalez Del-Pino, G, Ha, B.H, Park, E, Eck, M.J. | | Deposit date: | 2021-03-11 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Allosteric MEK inhibitors act on BRAF/MEK complexes to block MEK activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7B6F
 
 | | GSK3-beta in complex with compound (S)-5c | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3~{S})-3-[(7-chloranyl-9~{H}-pyrimido[4,5-b]indol-4-yl)-methyl-amino]piperidin-1-yl]propanenitrile, Glycogen synthase kinase-3 beta, ... | | Authors: | Tesch, R, Andreev, S, Koch, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | GSK3-beta in complex with compound (S)-5c
To be published
|
|
8SIX
 
 | | Structure of Compound 13 bound to the CHK1 10-point mutant | | Descriptor: | (1S)-N-(7-chloro-6-{4-[(3R,4R)-4-hydroxy-3-methyloxolan-3-yl]piperazin-1-yl}isoquinolin-3-yl)-6-oxaspiro[2.5]octane-1-carboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2023-04-17 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of MK-1468: A Potent, Kinome-Selective, Brain-Penetrant Amidoisoquinoline LRRK2 Inhibitor for the Potential Treatment of Parkinson's Disease.
J.Med.Chem., 66, 2023
|
|
7M1V
 
 | | Structure of Zika virus NS2b-NS3 protease mutant binding the compound NSC86314 in the super-open conformation | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-{2,4-diamino-5-[2-(4-{[(2E)-1,3-thiazolidin-2-ylidene]sulfamoyl}phenyl)hydrazinyl]phenyl}hydrazinyl)-N-[(2S)-1,3-thiazolidin-2-yl]benzene-1-sulfonamide, CHLORIDE ION, ... | | Authors: | Aleshin, A.E, Shiryaev, S.A, Liddington, R.C. | | Deposit date: | 2021-03-15 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | To be provided
To Be Published
|
|
8SUU
 
 | | Crystal structure of YisK from Bacillus subtilis in apo form | | Descriptor: | Fumarylacetoacetate hydrolase family protein, MANGANESE (II) ION | | Authors: | Krieger, I.V, Chemelewski, V, Guo, T, Sperber, A, Herman, J, Sacchettini, J.C. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Bacillus subtilis YisK possesses oxaloacetate decarboxylase activity and exhibits Mbl-dependent localization.
J.Bacteriol., 206, 2024
|
|
7M0Z
 
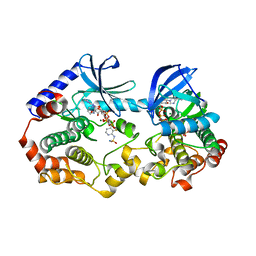 | | Crystal structure of the BRAF:MEK1 kinases in complex with AMPPNP and CH5126766 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, N-(3-fluoro-4-{[4-methyl-2-oxo-7-(pyrimidin-2-yloxy)-2H-chromen-3-yl]methyl}pyridin-2-yl)-N'-methylsulfuric diamide, ... | | Authors: | Li, K, Gonzalez Del-Pino, G, Ha, B.H, Park, E, Eck, M.J. | | Deposit date: | 2021-03-11 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Allosteric MEK inhibitors act on BRAF/MEK complexes to block MEK activation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ATS
 
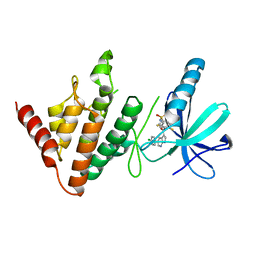 | | The LIMK1 Kinase Domain Bound To LIJTF500127 | | Descriptor: | LIM domain kinase 1, N-[3-[5-(4-Chlorophenyl)-1H-pyrrolo[2,3-b]pyridine-3-carbonyl]-2,4-difluorophenyl]benzenesulfonamide | | Authors: | Mathea, S, Chatterjee, D, Preuss, F, Yamamoto, S, Tawada, M, Nomura, I, Takagi, T, Ahmed, M, Little, W, Mueller-Knapp, S, Knapp, S. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The LIMK1 Kinase Domain Bound To LIJTF500127
To Be Published
|
|
7ZUO
 
 | |
6CPF
 
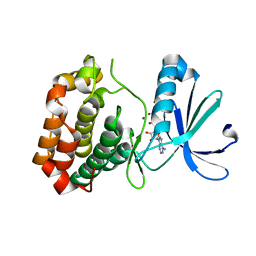 | | Structure of dephosphorylated Aurora A (122-403) bound to AMPPCP in an active conformation | | Descriptor: | Aurora kinase A, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Otten, R, Zorba, A, Padua, R.A.P, Kern, D. | | Deposit date: | 2018-03-13 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dynamics of human protein kinase Aurora A linked to drug selectivity.
Elife, 7, 2018
|
|
7ZVZ
 
 | |
7QEM
 
 | | bacterial IMPDH chimera | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Labesse, G, Gelin, M, Gedeon, A, Haouz, A, Munier-Lehmann, H. | | Deposit date: | 2021-12-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Insight into the role of the Bateman domain at the molecular and physiological levels through engineered IMP dehydrogenases.
Protein Sci., 32, 2023
|
|
6CW3
 
 | | Crystal structure of a yeast SAGA transcriptional coactivator Ada2/Gcn5 HAT subcomplex, crystal form 2 | | Descriptor: | Histone acetyltransferase GCN5, Transcriptional adapter 2, ZINC ION, ... | | Authors: | Sun, J, Paduch, M, Kim, S.A, Kramer, R.M, Barrios, A.F, Lu, V, Luke, J, Usatyuk, S, Kossiakoff, A.A, Tan, S. | | Deposit date: | 2018-03-29 | | Release date: | 2018-09-19 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7QQB
 
 | | Crystal structure of the envelope glycoprotein complex of Puumala virus in complex with the scFv fragment of the broadly neutralizing human antibody ADI-42898 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope polyprotein, GLYCEROL, ... | | Authors: | Serris, A, Rey, F.A, Guardado-Calvo, P. | | Deposit date: | 2022-01-07 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and mechanistic basis of neutralization by a pan-hantavirus protective antibody.
Sci Transl Med, 15, 2023
|
|
7AUC
 
 | |
7ZMY
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the ground state at pH 8.2 in the presence of sodium at 100K | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, OLEIC ACID, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZN0
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the M state at pH 8.2 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7QRL
 
 | |
7AV9
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2 | | Descriptor: | 1,2-ETHANEDIOL, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group C2
To Be Published
|
|
7ZN3
 
 | | Crystal structure of the light-driven inward proton pump xenorhodopsin BcXeR in the L state at pH 8.2 in the presence of sodium at 100K | | Descriptor: | EICOSANE, OLEIC ACID, PHOSPHATE ION, ... | | Authors: | Kovalev, K, Tsybrov, F, Alekseev, A, Bourenkov, G, Gordeliy, V. | | Deposit date: | 2022-04-20 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanisms of inward transmembrane proton translocation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7QVD
 
 | |
7AW6
 
 | | The extracellular region of CD33 with bound sialoside analogue P22 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-aminoethyl 5-{[(4-cyclohexyl-1H-1,2,3-triazol-1-yl)acetyl]amino}-3,5,9-trideoxy-9-[(4-hydroxy-3,5-dimethylbenzene-1-carbonyl)amino]-D-glycero-alpha-D-galacto-non-2-ulopyranonosyl-(2->6)-beta-D-galactopyranosyl-(1->4)-beta-D-glucopyranoside, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Thompson, A.P, Arrowsmith, C.H, Edwards, A.M, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2020-11-06 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The extracellular region of CD33 with bound sialoside analogue P22
To Be Published
|
|
7ZU4
 
 | |
