1PGR
 
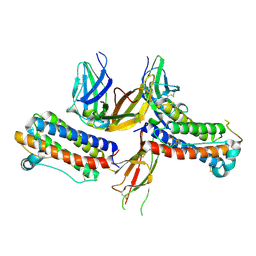 | | 2:2 COMPLEX OF G-CSF WITH ITS RECEPTOR | | Descriptor: | PROTEIN (G-CSF RECEPTOR), PROTEIN (GRANULOCYTE COLONY-STIMULATING FACTOR) | | Authors: | Aritomi, M, Kunishima, N, Okamoto, T, Kuroki, R, Ota, Y, Morikawa, K. | | Deposit date: | 1999-03-08 | | Release date: | 2000-03-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Atomic structure of the GCSF-receptor complex showing a new cytokine-receptor recognition scheme.
Nature, 401, 1999
|
|
1PIW
 
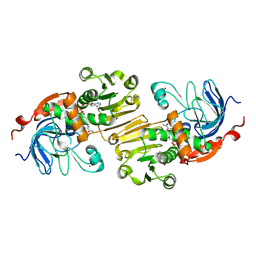 | | APO AND HOLO STRUCTURES OF AN NADP(H)-DEPENDENT CINNAMYL ALCOHOL DEHYDROGENASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | Hypothetical zinc-type alcohol dehydrogenase-like protein in PRE5-FET4 intergenic region, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Valencia, E, Larroy, C, Ochoa, W.F, Pares, X, Fita, I, Biosca, J.A. | | Deposit date: | 2003-05-30 | | Release date: | 2004-08-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Apo and Holo Structures of an NADP(H)-dependent Cinnamyl Alcohol Dehydrogenase from Saccharomyces cerevisiae
J.Mol.Biol., 341, 2004
|
|
1PJP
 
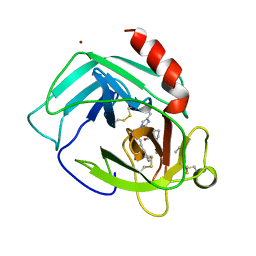 | | THE 2.2 A CRYSTAL STRUCTURE OF HUMAN CHYMASE IN COMPLEX WITH SUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYLKETONE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Chymase, SUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYLKETONE INHIBITOR, ... | | Authors: | Pereira, P.J.B, Wang, Z.M, Rubin, H, Huber, R, Bode, W, Schechter, N.M, Strobl, S. | | Deposit date: | 1998-09-07 | | Release date: | 1999-03-02 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A crystal structure of human chymase in complex with succinyl-Ala-Ala-Pro-Phe-chloromethylketone: structural explanation for its dipeptidyl carboxypeptidase specificity.
J.Mol.Biol., 286, 1999
|
|
1PJN
 
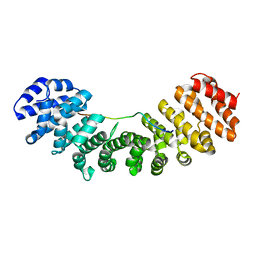 | | Mouse Importin alpha-bipartite NLS N1N2 from Xenopus laevis phosphoprotein Complex | | Descriptor: | Histone-binding protein N1/N2, Importin alpha-2 subunit | | Authors: | Fontes, M.R.M, Teh, T, Jans, D, Brinkworth, R.I, Kobe, B. | | Deposit date: | 2003-06-03 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-alpha
J.Biol.Chem., 278, 2003
|
|
1PJM
 
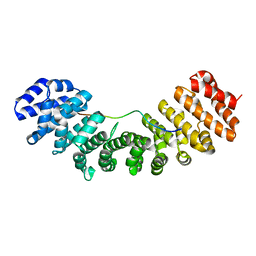 | | Mouse Importin alpha-bipartite NLS from human retinoblastoma protein Complex | | Descriptor: | Importin alpha-2 subunit, Retinoblastoma-associated protein | | Authors: | Fontes, M.R.M, Teh, T, Jans, D, Brinkworth, R.I, Kobe, B. | | Deposit date: | 2003-06-03 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-alpha
J.Biol.Chem., 278, 2003
|
|
1PMY
 
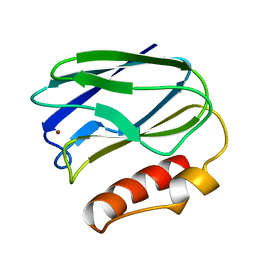 | | REFINED CRYSTAL STRUCTURE OF PSEUDOAZURIN FROM METHYLOBACTERIUM EXTORQUENS AM1 AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Inoue, T, Kai, Y, Harada, S, Kasai, N, Ohshiro, Y, Suzuki, S, Kohzuma, T, Tobari, J. | | Deposit date: | 1994-01-28 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Refined crystal structure of pseudoazurin from Methylobacterium extorquens AM1 at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1PNO
 
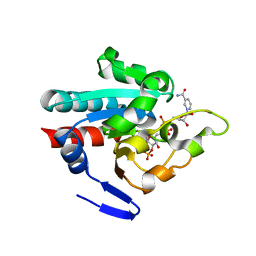 | | Crystal structure of R. rubrum transhydrogenase domain III bound to NADP | | Descriptor: | NAD(P) transhydrogenase subunit beta, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sundaresan, V, Yamaguchi, M, Chartron, J, Stout, C.D. | | Deposit date: | 2003-06-12 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Change in the NADP(H) Binding Domain of Transhydrogenase Defines Four States
Biochemistry, 42, 2003
|
|
1PNF
 
 | | PNGASE F COMPLEX WITH DI-N-ACETYLCHITOBIOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, PEPTIDE-N(4)-(N-ACETYL-BETA-D-GLUCOSAMINYL)ASPARAGINE AMIDASE F, SULFATE ION | | Authors: | Van Roey, P, Kuhn, P. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active site and oligosaccharide recognition residues of peptide-N4-(N-acetyl-beta-D-glucosaminyl)asparagine amidase F.
J.Biol.Chem., 270, 1995
|
|
1PO2
 
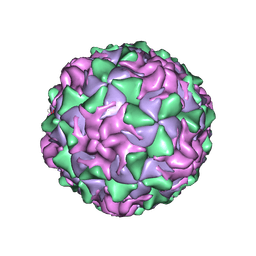 | | POLIOVIRUS (TYPE 1, MAHONEY) IN COMPLEX WITH R77975, AN INHIBITOR OF VIRAL REPLICATION | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE ETHYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 1 MAHONEY | | Authors: | Hiremath, C.N, Filman, D.J, Grant, R.A, Hogle, J.M. | | Deposit date: | 1997-01-08 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand-induced conformational changes in poliovirus-antiviral drug complexes.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1PS1
 
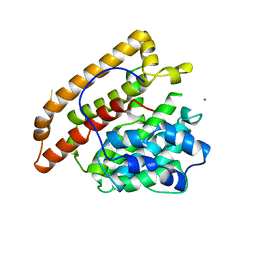 | | PENTALENENE SYNTHASE | | Descriptor: | PENTALENENE SYNTHASE, TRIMETHYL LEAD ION | | Authors: | Lesburg, C.A, Christianson, D.W. | | Deposit date: | 1997-03-23 | | Release date: | 1998-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of pentalenene synthase: mechanistic insights on terpenoid cyclization reactions in biology.
Science, 277, 1997
|
|
1PTX
 
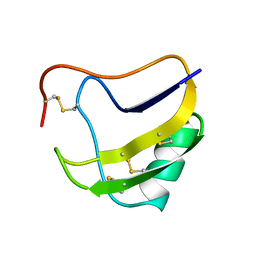 | |
1PSK
 
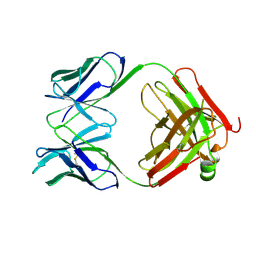 | |
1PTO
 
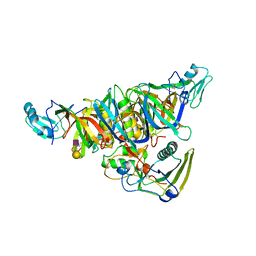 | |
1PVD
 
 | |
1PVX
 
 | | DO-1,4-BETA-XYLANASE, ROOM TEMPERATURE, PH 4.5 | | Descriptor: | PROTEIN (ENDO-1,4-BETA-XYLANASE) | | Authors: | Rajeshkumar, P, Eswaramoorthy, S, Vithayathil, P.J, Viswamitra, M.A. | | Deposit date: | 1998-10-20 | | Release date: | 1999-10-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The tertiary structure at 1.59 A resolution and the proposed amino acid sequence of a family-11 xylanase from the thermophilic fungus Paecilomyces varioti bainier.
J.Mol.Biol., 295, 2000
|
|
1PWP
 
 | | Crystal Structure of the Anthrax Lethal Factor complexed with Small Molecule Inhibitor NSC 12155 | | Descriptor: | Lethal factor, N,N'-BIS(4-AMINO-2-METHYLQUINOLIN-6-YL)UREA, ZINC ION | | Authors: | Wong, T.Y, Schwarzenbacher, R, Liddington, R.C. | | Deposit date: | 2003-07-02 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of small molecule inhibitors of anthrax lethal factor.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1PXC
 
 | | CRYSTAL STRUCTURES OF MUTANT PSEUDOMONAS AERUGINOSA P-HYDROXYBENZOATE HYDROXYLASE: THE TYR201PHE, TYR385PHE, AND ASN300ASP VARIANTS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Lah, M.S, Palfey, B.A, Schreuder, H.A, Ludwig, M.L. | | Deposit date: | 1994-09-27 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of mutant Pseudomonas aeruginosa p-hydroxybenzoate hydroxylases: the Tyr201Phe, Tyr385Phe, and Asn300Asp variants.
Biochemistry, 33, 1994
|
|
1Q07
 
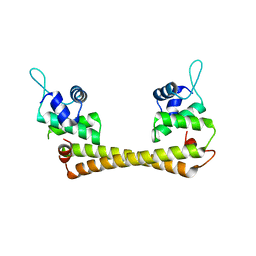 | | Crystal structure of the Au(I) form of E. coli CueR, a copper efflux regulator | | Descriptor: | GOLD ION, Transcriptional regulator cueR | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1PYT
 
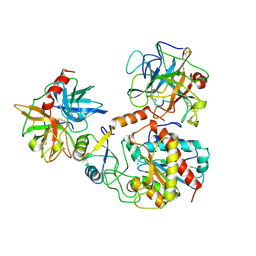 | | TERNARY COMPLEX OF PROCARBOXYPEPTIDASE A, PROPROTEINASE E, AND CHYMOTRYPSINOGEN C | | Descriptor: | CALCIUM ION, CHYMOTRYPSINOGEN C, PROCARBOXYPEPTIDASE A, ... | | Authors: | Gomis-Ruth, F.X, Gomez, M, Bode, W, Huber, R, Aviles, F.X. | | Deposit date: | 1995-06-21 | | Release date: | 1997-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The three-dimensional structure of the native ternary complex of bovine pancreatic procarboxypeptidase A with proproteinase E and chymotrypsinogen C.
EMBO J., 14, 1995
|
|
1PZC
 
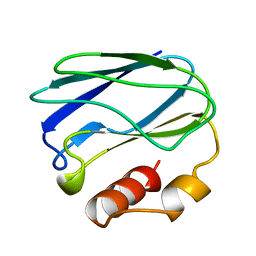 | | APO-PSEUDOAZURIN (METAL FREE PROTEIN) | | Descriptor: | PSEUDOAZURIN | | Authors: | Petratos, K. | | Deposit date: | 1995-02-22 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of apo-pseudoazurin from Alcaligenes faecalis S-6.
Febs Lett., 368, 1995
|
|
1Q1W
 
 | |
1Q2V
 
 | | Crystal structure of the chaperonin from Thermococcus strain KS-1 (nucleotide-free form) | | Descriptor: | SULFATE ION, Thermosome alpha subunit | | Authors: | Shomura, Y, Yoshida, T, Iizuka, R, Yohda, M, Maruyama, T, Miki, K. | | Deposit date: | 2003-07-26 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the Group II Chaperonin from Thermococcus strain KS-1: Steric Hindrance by the Substituted Amino Acid, and Inter-subunit Rearrangement between Two Crystal Forms.
J.Mol.Biol., 335, 2004
|
|
1MO0
 
 | | Structural Genomics Of Caenorhabditis Elegans: Triose Phosphate Isomerase | | Descriptor: | ACETATE ION, SULFATE ION, Triosephosphate isomerase | | Authors: | Symersky, J, Li, S, Finley, J, Liu, Z.-J, Qui, H, Luan, C.H, Carson, M, Tsao, J, Johnson, D, Lin, G, Zhao, J, Thomas, W, Nagy, L.A, Sha, B, DeLucas, L.J, Wang, B.-C, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2002-09-06 | | Release date: | 2002-09-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: triosephosphate isomerase
Proteins, 51, 2003
|
|
1MOF
 
 | | COAT PROTEIN | | Descriptor: | CHLORIDE ION, MOLONEY MURINE LEUKEMIA VIRUS P15 | | Authors: | Fass, D, Harrison, S.C, Kim, P.S. | | Deposit date: | 1996-04-02 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Retrovirus envelope domain at 1.7 angstrom resolution.
Nat.Struct.Biol., 3, 1996
|
|
1MPD
 
 | |
