3V6Z
 
 | | Crystal Structure of Hepatitis B Virus e-antigen | | Descriptor: | Fab e6 Heavy Chain, Fab e6 Light Chain, e-antigen | | Authors: | Dimattia, M.A, Watts, N.R, Stahl, S.J, Grimes, J.M, Steven, A.C, Stuart, D.I, Wingfield, P.T. | | Deposit date: | 2011-12-20 | | Release date: | 2013-02-06 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Antigenic switching of hepatitis B virus by alternative dimerization of the capsid protein.
Structure, 21, 2013
|
|
2C9W
 
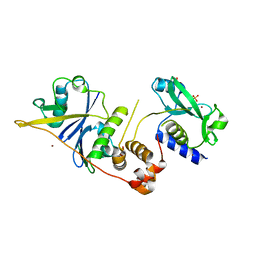 | | CRYSTAL STRUCTURE OF SOCS-2 IN COMPLEX WITH ELONGIN-B AND ELONGIN-C AT 1.9A RESOLUTION | | Descriptor: | NICKEL (II) ION, SULFATE ION, SUPPRESSOR OF CYTOKINE SIGNALING 2, ... | | Authors: | Debreczeni, J.E, Bullock, A, Amos, A, Savitsky, P, Barr, A, Burgess, N, Sundstrom, M, Weigelt, J, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2005-12-14 | | Release date: | 2006-02-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SOCS2-elongin C-elongin B complex defines a prototypical SOCS box ubiquitin ligase.
Proc. Natl. Acad. Sci. U.S.A., 103, 2006
|
|
4M2U
 
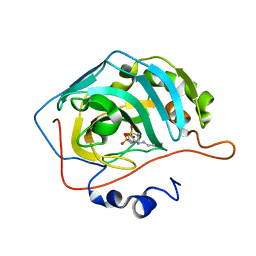 | | Carbonic Anhydrase II in complex with Dorzolamide | | Descriptor: | (4S-TRANS)-4-(ETHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Pinard, M.P, Boone, C.D, Rife, B.D, Supuran, C.T, Mckenna, R. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural study of interaction between brinzolamide and dorzolamide inhibition of human carbonic anhydrases.
Bioorg.Med.Chem., 21, 2013
|
|
1XDM
 
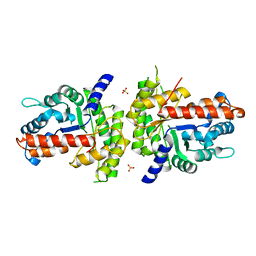 | | Structure of human aldolase B associated with hereditary fructose intolerance (A149P), at 291K | | Descriptor: | Fructose-bisphosphate aldolase B, SULFATE ION | | Authors: | Malay, A.D, Allen, K.N, Tolan, D.R. | | Deposit date: | 2004-09-07 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the thermolabile mutant aldolase B, A149P: molecular basis of hereditary fructose intolerance.
J.Mol.Biol., 347, 2005
|
|
1XDL
 
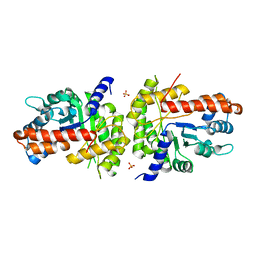 | | Structure of human aldolase B associated with hereditary fructose intolerance (A149P), at 277K | | Descriptor: | Fructose-bisphosphate aldolase B, SULFATE ION | | Authors: | Malay, A.D, Allen, K.N, Tolan, D.R. | | Deposit date: | 2004-09-07 | | Release date: | 2005-03-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the thermolabile mutant aldolase B, A149P: molecular basis of hereditary fructose intolerance.
J.Mol.Biol., 347, 2005
|
|
7XSV
 
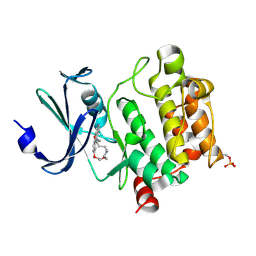 | | Crystal Structures of PIM1 in Complex with Macrocyclic Compound H3 | | Descriptor: | 8-Methyl-2,5,20-trioxa-8,13,17-triazatetracyclo[11.10.2.014,19.021,25]pentacosa-1(24),14(19),15,17,21(25),22-hexaene, Serine/threonine-protein kinase pim-1 | | Authors: | Shen, C, Xie, Y, Ren, X, Zhou, Y, Niu, H. | | Deposit date: | 2022-05-15 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Design, synthesis, and bioactivity evaluation of macrocyclic benzo[b]pyrido[4,3-e][1,4]oxazine derivatives as novel Pim-1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 72, 2022
|
|
1IWD
 
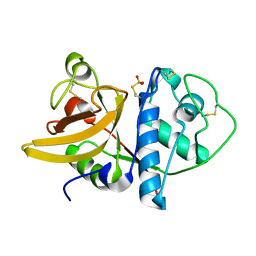 | |
1YWM
 
 | | Crystal structure of the N-terminal domain of group B Streptococcus alpha C protein | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, C protein alpha-antigen, GLYCEROL | | Authors: | Auperin, T.C, Bolduc, G.R, Baron, M.J, Heroux, A, Filman, D.J, Madoff, L.C, Hogle, J.M. | | Deposit date: | 2005-02-18 | | Release date: | 2005-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the N-terminal domain of the group B streptococcus alpha C protein.
J.Biol.Chem., 280, 2005
|
|
3H0Z
 
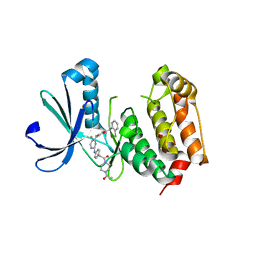 | | Aurora A in complex with a bisanilinopyrimidine | | Descriptor: | 4-{[2-({4-[2-(4-acetylpiperazin-1-yl)-2-oxoethyl]phenyl}amino)-5-fluoropyrimidin-4-yl]amino}-N-(2-chlorophenyl)benzamide, Serine/threonine-protein kinase 6 | | Authors: | Wiesmann, C, Ultsch, M.H, Cochran, A.G. | | Deposit date: | 2009-04-10 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | A class of 2,4-bisanilinopyrimidine Aurora A inhibitors with unusually high selectivity against Aurora B.
J.Med.Chem., 52, 2009
|
|
4AF3
 
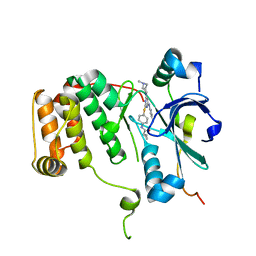 | | Human Aurora B Kinase in complex with INCENP and VX-680 | | Descriptor: | AURORA KINASE B, CYCLOPROPANECARBOXYLIC ACID {4-[4-(4-METHYL-PIPERAZIN-1-YL)-6-(5-METHYL-2H-PYRAZOL-3-YLAMINO)-PYRIMIDIN-2-YLSULFANYL]-PHENYL}-AMIDE, INNER CENTROMERE PROTEIN | | Authors: | Elkins, J.M, Vollmar, M, Wang, J, Picaud, S, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Knapp, S. | | Deposit date: | 2012-01-16 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of Human Aurora B in Complex with Incenp and Vx-680.
J.Med.Chem., 55, 2012
|
|
1DYP
 
 | | 1,3-ALPHA-1,4-BETA-D-GALACTOSE-4-SULFATE-3,6-ANHYDRO-D-GALACTOSE 4 GALACTOHYDROLASE | | Descriptor: | CADMIUM ION, CHLORIDE ION, KAPPA-CARRAGEENASE | | Authors: | Michel, G, Chantalat, L, Dideberg, O. | | Deposit date: | 2000-02-04 | | Release date: | 2001-01-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The Kappa-Carrageenase of P. Carrageenovora Features a Tunnel-Shaped Active Site: A Novel Insight in the Evolution of Clan-B Glycoside Hydrolases
Structure, 9, 2001
|
|
3H10
 
 | | Aurora A inhibitor complex | | Descriptor: | 9-chloro-7-(2,6-difluorophenyl)-N-{4-[(4-methylpiperazin-1-yl)carbonyl]phenyl}-5H-pyrimido[5,4-d][2]benzazepin-2-amine, Serine/threonine-protein kinase 6 | | Authors: | Wiesmann, C, Ultsch, M.H, Cochran, A.G. | | Deposit date: | 2009-04-10 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A class of 2,4-bisanilinopyrimidine Aurora A inhibitors with unusually high selectivity against Aurora B.
J.Med.Chem., 52, 2009
|
|
4GD3
 
 | | Structure of E. coli hydrogenase-1 in complex with cytochrome b | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2012-07-31 | | Release date: | 2013-01-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the O(2)-Tolerant Membrane-Bound Hydrogenase 1 from Escherichia coli in Complex with Its Cognate Cytochrome b.
Structure, 21, 2013
|
|
2QC5
 
 | | Streptogramin B lyase structure | | Descriptor: | IODIDE ION, Streptogramin B lactonase | | Authors: | Lipka, M, Bochtler, M. | | Deposit date: | 2007-06-19 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and mechanism of the Staphylococcus cohnii virginiamycin B lyase (Vgb).
Biochemistry, 47, 2008
|
|
2KHK
 
 | | NMR solution structure of the b30-82 domain of subunit b of Escherichia coli F1FO ATP synthase | | Descriptor: | ATP synthase subunit b | | Authors: | Priya, R, Biukovic, G, Gayen, S, Vivekanandan, S, Gruber, G. | | Deposit date: | 2009-04-08 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure, determined by nuclear magnetic resonance, of the b30-82 domain of subunit b of Escherichia coli F1Fo ATP synthase
J.Bacteriol., 191, 2009
|
|
1QDQ
 
 | | X-RAY CRYSTAL STRUCTURE OF BOVINE CATHEPSIN B-CA074 COMPLEX | | Descriptor: | CATHEPSIN B, [PROPYLAMINO-3-HYDROXY-BUTAN-1,4-DIONYL]-ISOLEUCYL-PROLINE | | Authors: | Yamamoto, A. | | Deposit date: | 1999-07-10 | | Release date: | 2000-07-10 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Substrate specificity of bovine cathepsin B and its inhibition by CA074, based on crystal structure refinement of the complex.
J.Biochem.(Tokyo), 127, 2000
|
|
3HCH
 
 | | Structure of the C-terminal domain (MsrB) of Neisseria meningitidis PilB (complex with substrate) | | Descriptor: | (2S)-2-(acetylamino)-N-methyl-4-[(R)-methylsulfinyl]butanamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRIC ACID, ... | | Authors: | Ranaivoson, F.M, Kauffmann, B, Favier, F. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methionine sulfoxide reductase B displays a high level of flexibility.
J.Mol.Biol., 394, 2009
|
|
2JG8
 
 | | Crystallographic structure of human C1q globular heads complexed to phosphatidyl-serine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement C1q subcomponent subunit A, ... | | Authors: | Paidassi, H, Tacnet-Delorme, P, Garlatti, V, Darnault, C, Ghebrehiwet, B, Gaboriaud, C, Arlaud, G.J, Frachet, P. | | Deposit date: | 2007-02-09 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | C1Q Binds Phosphatidylserine and Likely Acts as a Multiligand-Bridging Molecule in Apoptotic Cell Recognition.
J.Immunol., 180, 2008
|
|
3TWI
 
 | | Variable Lymphocyte Receptor Recognition of the Immunodominant Glycoprotein of Bacillus anthracis Spores | | Descriptor: | BclA protein, GLYCEROL, Variable lymphocyte receptor B | | Authors: | Kirchdoerfer, R.N, Herrin, B.R, Han, B.W, Turnbough Jr, C.L, Cooper, M.D, Wilson, I.A. | | Deposit date: | 2011-09-21 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Variable Lymphocyte Receptor Recognition of the Immunodominant Glycoprotein of Bacillus anthracis Spores.
Structure, 20, 2012
|
|
2JKH
 
 | | Factor Xa - cation inhibitor complex | | Descriptor: | 3-[(3~{a}~{S},4~{R},8~{a}~{S},8~{b}~{R})-4-[5-(5-chloranylthiophen-2-yl)-1,2-oxazol-3-yl]-1,3-bis(oxidanylidene)-4,6,7,8,8~{a},8~{b}-hexahydro-3~{a}~{H}-pyrrolo[3,4-a]pyrrolizin-2-yl]propyl-trimethyl-azanium, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Salonen, L.M, Bucher, C, Banner, D.W, Benz, J, Haap, W, Mary, J.L, Schweizer, W.B, Seiler, P, Kuster, O, Diederich, F. | | Deposit date: | 2008-08-28 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Cation-Pi Interactions at the Active Site of Factor Xa: Dramatic Enhancement Upon Stepwise N-Alkylation of Ammonium Ions.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2LCT
 
 | |
1WC2
 
 | | Beta-1,4-D-endoglucanase Cel45A from blue mussel Mytilus edulis at 1.2A | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, ENDOGLUCANASE | | Authors: | Jakobsson, E, Mahdi, S, Kleywegt, G.J, Stahlberg, J. | | Deposit date: | 2004-11-08 | | Release date: | 2006-05-24 | | Last modified: | 2021-12-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Glucomannan and beta-glucan degradation by Mytilus edulis Cel45A: Crystal structure and activity comparison with GH45 subfamily A, B and C.
Carbohydr Polym, 277, 2022
|
|
4G93
 
 | |
4O8U
 
 | | Structure of PF2046 | | Descriptor: | Uncharacterized protein PF2046 | | Authors: | Su, J, Liu, Z.-J. | | Deposit date: | 2013-12-30 | | Release date: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (2.345 Å) | | Cite: | Crystal structure of a novel non-Pfam protein PF2046 solved using low resolution B-factor sharpening and multi-crystal averaging methods
Protein Cell, 1, 2010
|
|
4OLR
 
 | | [Leu-5]-Enkephalin mutant - YVVFV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, [Leu-5]-Enkephalin mutant - YVVFV | | Authors: | Sangwan, S, Eisenberg, D, Sawaya, M.R, Do, T.D, Bowers, M.T, Lapointe, N.E, Teplow, D.B, Feinstein, S.C. | | Deposit date: | 2014-01-24 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Factors that drive Peptide assembly from native to amyloid structures: experimental and theoretical analysis of [leu-5]-enkephalin mutants.
J.Phys.Chem.B, 118, 2014
|
|
