1ZLR
 
 | | Factor XI catalytic domain complexed with 2-guanidino-1-(4-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)phenyl)ethyl nicotinate | | Descriptor: | (1R)-2-{[AMINO(IMINO)METHYL]AMINO}-1-{4-[(4R)-4-(HYDROXYMETHYL)-1,3,2-DIOXABOROLAN-2-YL]PHENYL}ETHYL NICOTINATE, Coagulation factor XI, GLYCEROL, ... | | Authors: | Lazarova, T.I, Jin, L, Rynkiewicz, M, Gorga, J.C, Bibbins, F, Meyers, H.V, Babine, R, Strickler, J. | | Deposit date: | 2005-05-09 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthesis and in vitro biological evaluation of aryl boronic acids as potential inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1L6Y
 
 | | Crystal Structure of Porphobilinogen Synthase Complexed with the Inhibitor 4-Oxosebacic Acid | | Descriptor: | 4-OXODECANEDIOIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Jaffe, E.K, Kervinen, J, Martins, J, Stauffer, F, Neier, R, Wlodawer, A, Zdanov, A. | | Deposit date: | 2002-03-14 | | Release date: | 2002-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Species-Specific Inhibition of Porphobilinogen Synthase by 4-Oxosebacic Acid
J.Biol.Chem., 277, 2002
|
|
1KBO
 
 | | Complex of Human recombinant NAD(P)H:Quinone Oxide reductase type 1 with 5-methoxy-1,2-dimethyl-3-(phenoxymethyl)indole-4,7-dione (ES1340) | | Descriptor: | 5-METHOXY-1,2-DIMETHYL-3-(PHENOXYMETHYL)INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Faig, M, Bianchet, M.A, Amzel, L.M. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of a mechanism-based inhibitor of NAD(P)H:quinone oxidoreductase 1 by biochemical, X-ray crystallographic, and mass spectrometric approaches.
Biochemistry, 40, 2001
|
|
2QEX
 
 | |
1LJH
 
 | |
2ZMN
 
 | | Crystal Structure of basic winged bean lectin in complex with Gal-alpha- 1,6 Glc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Basic agglutinin, CALCIUM ION, ... | | Authors: | Kulkarni, K.A, Katiyar, S, Surolia, A, Vijayan, M, Suguna, K. | | Deposit date: | 2008-04-19 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and sugar-specificity of basic winged-bean lectin: structures of new disaccharide complexes and a comparative study with other known disaccharide complexes of the lectin.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2X2I
 
 | | Crystal structure of the Gracilariopsis lemaneiformis alpha-1,4- glucan lyase with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, GLYCEROL | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
1V3L
 
 | | Crystal structure of F283L mutant cyclodextrin glycosyltransferase complexed with a pseudo-tetraose derived from acarbose | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-beta-D-galactopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2003-11-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of Phe283 in enzymatic reaction of cyclodextrin glycosyltransferase from alkalophilic Bacillus sp.1011: Substrate binding and arrangement of the catalytic site
PROTEIN SCI., 13, 2004
|
|
2EI4
 
 | | Trimeric complex of archaerhodopsin-2 | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Archaerhodopsin-2, BACTERIORUBERIN, ... | | Authors: | Kouyama, T. | | Deposit date: | 2007-03-11 | | Release date: | 2008-01-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural role of bacterioruberin in the trimeric structure of archaerhodopsin-2
J.Mol.Biol., 375, 2008
|
|
2ZNN
 
 | | Human PPAR alpha ligand binding domain in complex with a synthetic agonist TIPP703 | | Descriptor: | (2S)-2-(4-propoxy-3-{[({4-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]phenyl}carbonyl)amino]methyl}benzyl)butanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Oyama, T, Toyota, K, Kasuga, J, Miyachi, H, Morikawa, K. | | Deposit date: | 2008-04-30 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Adaptability and selectivity of human peroxisome proliferator-activated receptor (PPAR) pan agonists revealed from crystal structures
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2Z55
 
 | |
1ULV
 
 | | Crystal Structure of Glucodextranase Complexed with Acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, glucodextranase | | Authors: | Mizuno, M, Tonozuka, T, Suzuki, S, Uotsu-Tomita, R, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2003-09-16 | | Release date: | 2003-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural insights into substrate specificity and function of glucodextranase
J.Biol.Chem., 279, 2004
|
|
1KXA
 
 | |
1KFI
 
 | | Crystal Structure of the Exocytosis-Sensitive Phosphoprotein, pp63/Parafusin (phosphoglucomutase) from Paramecium | | Descriptor: | SULFATE ION, ZINC ION, phosphoglucomutase 1 | | Authors: | Mueller, S, Diederichs, K, Breed, J, Kissmehl, R, Hauser, K, Plattner, H, Welte, W. | | Deposit date: | 2001-11-21 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure analysis of the exocytosis-sensitive phosphoprotein, pp63/parafusin (phosphoglucomutase), from Paramecium reveals significant conformational variability.
J.Mol.Biol., 315, 2002
|
|
1KX8
 
 | | Antennal Chemosensory Protein A6 from Mamestra brassicae, tetragonal form | | Descriptor: | CHEMOSENSORY PROTEIN A6 | | Authors: | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray Structure and Ligand Binding Study of a Chemosensory Protein
J.Biol.Chem., 277, 2002
|
|
1LJG
 
 | |
2WSP
 
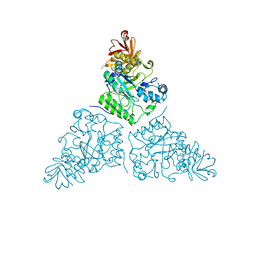 | | Thermotoga maritima alpha-L-fucosynthase, TmD224G, in complex with alpha-L-Fuc-(1-2)-beta-L-Fuc-N3 | | Descriptor: | ALPHA-L-FUCOSIDASE, PUTATIVE, alpha-L-fucopyranose-(1-2)-beta-L-fucosyl-azide | | Authors: | Sulzenbacher, G, Lipski, A, Cobucci-Ponzano, B, Conte, F, Bedini, E, Corsaro, M.M, Parrilli, M, Dal Piaz, F, Lepore, L, Rossi, M, Moracci, M. | | Deposit date: | 2009-09-08 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Beta-Glycosyl Azides as Substrates for Alpha-Glycosynthases: Preparation of Novel Efficient Alpha-L-Fucosynthases
Chem.Biol., 16, 2009
|
|
1PPH
 
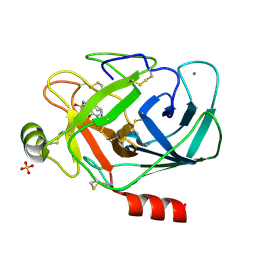 | | GEOMETRY OF BINDING OF THE NALPHA-TOSYLATED PIPERIDIDES OF M-AMIDINO-, P-AMIDINO-AND P-GUANIDINO PHENYLALANINE TO THROMBIN AND TRYPSIN: X-RAY CRYSTAL STRUCTURES OF THEIR TRYPSIN COMPLEXES AND MODELING OF THEIR THROMBIN COMPLEXES | | Descriptor: | 3-[(2S)-2-{[(4-methylphenyl)sulfonyl]amino}-3-oxo-3-piperidin-1-ylpropyl]benzenecarboximidamide, CALCIUM ION, SULFATE ION, ... | | Authors: | Bode, W, Turk, D. | | Deposit date: | 1991-10-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Geometry of binding of the N alpha-tosylated piperidides of m-amidino-, p-amidino- and p-guanidino phenylalanine to thrombin and trypsin. X-ray crystal structures of their trypsin complexes and modeling of their thrombin complexes.
FEBS Lett., 287, 1991
|
|
2ESU
 
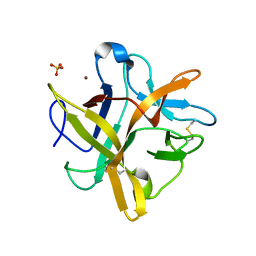 | | Crystal structure of Asn to Gln mutant of Winged Bean Chymotrypsin Inhibitor protein | | Descriptor: | Chymotrypsin inhibitor 3, NICKEL (II) ION, SULFATE ION | | Authors: | Dattagupta, J.K, Sen, U, Dasgupta, J, Khamrui, S. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Spacer Asn Determines the Fate of Kunitz (STI) Inhibitors, as Revealed by Structural and Biochemical Studies on WCI Mutants.
Biochemistry, 45, 2006
|
|
1LC0
 
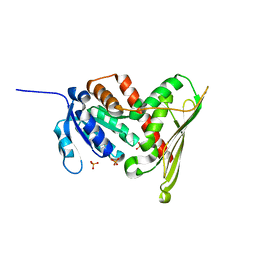 | | Structure of Biliverdin Reductase and the Enzyme-NADH Complex | | Descriptor: | Biliverdin Reductase A, PHOSPHATE ION | | Authors: | Whitby, F.G, Phillips, J.D, Hill, C.P, McCoubrey, W, Maines, M.D. | | Deposit date: | 2002-04-04 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a biliverdin IXalpha reductase enzyme-cofactor complex.
J.Mol.Biol., 319, 2002
|
|
1JTW
 
 | |
3WB4
 
 | | Crystal Structure of beta secetase in complex with 2-amino-3,6-dimethyl-6-(2-phenylethyl)-3,4,5,6-tetrahydropyrimidin-4-one | | Descriptor: | (6R)-2-amino-3,6-dimethyl-6-(2-phenylethyl)-5,6-dihydropyrimidin-4(3H)-one, Beta-secretase 1, GLYCEROL, ... | | Authors: | Yonezawa, S, Fujiwara, K, Yamamoto, T, Hattori, K, Yamakawa, H, Muto, C, Hosono, M, Tanaka, Y, Nakano, T, Takemoto, H, Arisawa, M, Shuto, S. | | Deposit date: | 2013-05-13 | | Release date: | 2013-10-02 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Conformational restriction approach to beta-secretase (BACE1) inhibitors III: Effective investigation of the binding mode by combinational use of X-ray analysis, isothermal titration calorimetry and theoretical calculations
Bioorg.Med.Chem., 21, 2013
|
|
1ZLX
 
 | | The apo structure of human glycinamide ribonucleotide transformylase | | Descriptor: | GLYCEROL, Phosphoribosylglycinamide formyltransferase | | Authors: | Dahms, T.E, Sainz, G, Giroux, E.L, Caperelli, C.A, Smith, J.L. | | Deposit date: | 2005-05-09 | | Release date: | 2005-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The apo and ternary complex structures of a chemotherapeutic target: human glycinamide ribonucleotide transformylase.
Biochemistry, 44, 2005
|
|
1JTU
 
 | | E. coli Thymidylate Synthase in a Complex with dUMP and LY338913, A Polyglutamylated Pyrrolo(2,3-d)pyrimidine-based Antifolate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-{4-[4-(4-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-4-CARBOXY-BUTYRYLAMIN O)-4-CARBOXY-BUTYRYLAMINO}-PENTANEDIOIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Sayre, P.H, Finer-Moore, J.S, Fritz, T.A, Biermann, D, Gates, S.B, MacKellar, W.C, Patel, V.F, Stroud, R.M. | | Deposit date: | 2001-08-22 | | Release date: | 2001-09-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multi-targeted antifolates aimed at avoiding drug resistance form covalent closed inhibitory complexes with human and Escherichia coli thymidylate synthases.
J.Mol.Biol., 313, 2001
|
|
2EUD
 
 | | Structures of Yeast Ribonucleotide Reductase I complexed with Ligands and Subunit Peptides | | Descriptor: | GEMCITABINE DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Dealwis, C, Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J. | | Deposit date: | 2005-10-28 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of eukaryotic ribonucleotide reductase I define gemcitabine diphosphate binding and subunit assembly.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
