3QMR
 
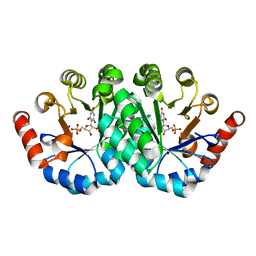 | | Crystal structure of the mutant R160A,V182A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-02-05 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3213 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
4HJ3
 
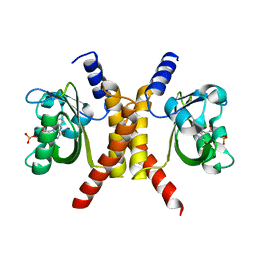 | |
3QMT
 
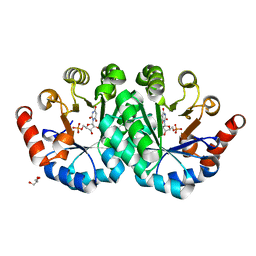 | | Crystal structure of the mutant V182A,Y206F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-02-05 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3202 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
1TRB
 
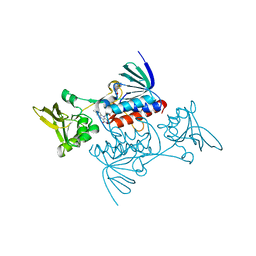 | |
3QRF
 
 | | Structure of a domain-swapped FOXP3 dimer | | Descriptor: | Forkhead box protein P3, MAGNESIUM ION, Nuclear factor of activated T-cells, ... | | Authors: | Bandukwala, H.S, Wu, Y, Feurer, M, Chen, Y, Barbosa, B, Ghosh, S, Stroud, J.C, Benoist, C, Mathis, D, Rao, A, Chen, L. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a Domain-Swapped FOXP3 Dimer on DNA and Its Function in Regulatory T Cells.
Immunity, 34, 2011
|
|
3SBM
 
 | | Trans-acting transferase from Disorazole synthase in complex with Acetate | | Descriptor: | ACETATE ION, DisD protein, HEXAETHYLENE GLYCOL | | Authors: | Khosla, C, Mathews, I.I, Wong, F.T, Jin, X. | | Deposit date: | 2011-06-06 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and Mechanism of the trans-Acting Acyltransferase from the Disorazole Synthase.
Biochemistry, 50, 2011
|
|
4I16
 
 | | Crystal structure of CARMA1 CARD | | Descriptor: | Caspase recruitment domain-containing protein 11, SULFATE ION | | Authors: | Li, S, Yang, X, Shen, Y. | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Structural insights into the assembly of CARMA1 and BCL10
Plos One, 7, 2012
|
|
8QLP
 
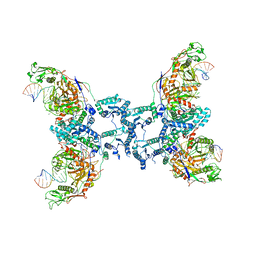 | | CryoEM structure of the RNA/DNA bound SPARTA (BabAgo/TIR-APAZ) tetrameric complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*AP*TP*TP*AP*GP*AP*GP*CP*CP*GP*TP*C)-3'), MAGNESIUM ION, RNA (5'-R(*AP*UP*GP*AP*CP*GP*GP*CP*UP*CP*UP*AP*AP*UP*CP*UP*AP*UP*UP*AP*GP*U)-3'), ... | | Authors: | Finocchio, G, Koopal, B, Potocnik, A, Heijstek, C, Jinek, M, Swarts, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Target DNA-dependent activation mechanism of the prokaryotic immune system SPARTA.
Nucleic Acids Res., 52, 2024
|
|
5C7K
 
 | | Crystal structure BG505 SOSIP gp140 HIV-1 Env trimer bound to broadly neutralizing antibodies PGT128 and 8ANC195 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab 8ANC195 heavy chain, ... | | Authors: | Kong, L, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2015-06-24 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.6019 Å) | | Cite: | Complete epitopes for vaccine design derived from a crystal structure of the broadly neutralizing antibodies PGT128 and 8ANC195 in complex with an HIV-1 Env trimer.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7MTG
 
 | | Structure of the adeno-associated virus 9 capsid at pH 6.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
6AVG
 
 | | Crystal structure of the KFJ37 TCR-NY-ESO-1-HLA-B*07:02 complex | | Descriptor: | ALA-PRO-ARG-GLY-PRO-HIS-GLY-GLY-ALA-ALA-SER-GLY-LEU, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gully, B.S, Gras, S, Rossjohn, J. | | Deposit date: | 2017-09-02 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Divergent T-cell receptor recognition modes of a HLA-I restricted extended tumour-associated peptide.
Nat Commun, 9, 2018
|
|
7MTP
 
 | | Structure of the adeno-associated virus 9 capsid at pH 5.5 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTW
 
 | | Structure of the adeno-associated virus 9 capsid at pH 4.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
8QLO
 
 | | CryoEM structure of the apo SPARTA (BabAgo/TIR-APAZ) complex | | Descriptor: | Short prokaryotic Argonaute, Toll/interleukin-1 receptor domain-containing protein | | Authors: | Finocchio, G, Koopal, B, Potocnik, A, Heijstek, C, Jinek, M, Swarts, D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Target DNA-dependent activation mechanism of the prokaryotic immune system SPARTA.
Nucleic Acids Res., 52, 2024
|
|
1P7H
 
 | | Structure of NFAT1 bound as a dimer to the HIV-1 LTR kB element | | Descriptor: | 5'-D(*AP*AP*TP*GP*GP*GP*GP*AP*CP*TP*TP*TP*CP*CP*A)-3', 5'-D(*TP*TP*TP*GP*GP*AP*AP*AP*GP*TP*CP*CP*CP*CP*A)-3', Nuclear factor of activated T-cells, ... | | Authors: | Giffin, M.J, Stroud, J.C, Bates, D.L, von Koenig, K.D, Hardin, J, Chen, L. | | Deposit date: | 2003-05-01 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of NFAT1 bound as a dimer to the HIV-1 LTR kappa B element
Nat.Struct.Biol., 10, 2003
|
|
6E3C
 
 | |
5OKE
 
 | | Conservatively refined structure of Gan1D-E170Q, a catalytic mutant of a putative 6-phospho-beta-galactosidase from Geobacillus stearothermophilus, in complex with cellobiose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2017-07-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural basis for enzyme bifunctionality - the case of Gan1D from Geobacillus stearothermophilus.
FEBS J., 284, 2017
|
|
6QXF
 
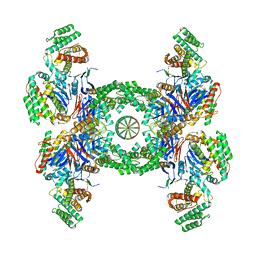 | | Cas1-Cas2-Csn2-DNA complex from the Type II-A CRISPR-Cas system | | Descriptor: | CALCIUM ION, CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, ... | | Authors: | Wilkinson, M, Drabavicius, G, Silanskas, A, Gasiunas, G, Siksnys, V, Wigley, D.B. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the DNA-Bound Spacer Capture Complex of a Type II CRISPR-Cas System.
Mol.Cell, 75, 2019
|
|
8QMF
 
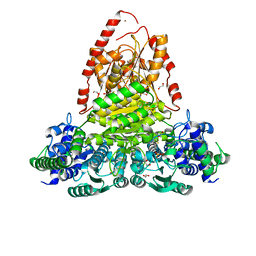 | | Transketolase from Vibrio vulnificus in complex with thiamin pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Ballut, L, Georges, R.N, Octobre, G, Charmantray, F, Doumeche, B. | | Deposit date: | 2023-09-22 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural determination and kinetic analysis of the transketolase from Vibrio vulnificus reveal unexpected cooperative behavior.
Protein Sci., 33, 2024
|
|
6E4H
 
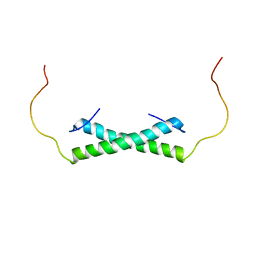 | | Solution NMR Structure of the Colied-coil PALB2 Homodimer | | Descriptor: | Partner and localizer of BRCA2 | | Authors: | Song, F, Li, M, Liu, G, Swapna, G.V.T, Xia, B, Bunting, S.F, Montelione, G.T. | | Deposit date: | 2018-07-17 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Antiparallel Coiled-Coil Interactions Mediate the Homodimerization of the DNA Damage-Repair Protein PALB2.
Biochemistry, 57, 2018
|
|
5ODR
 
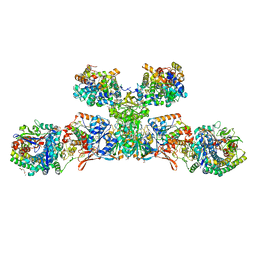 | | Heterodisulfide reductase / [NiFe]-hydrogenase complex from Methanothermococcus thermolithotrophicus soaked with heterodisulfide for 2 minutes. | | Descriptor: | 1-THIOETHANESULFONIC ACID, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETATE ION, ... | | Authors: | Wagner, T, Koch, J, Ermler, U, Shima, S. | | Deposit date: | 2017-07-06 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction.
Science, 357, 2017
|
|
8G5C
 
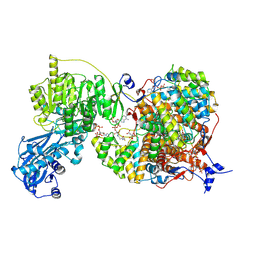 | |
5XKM
 
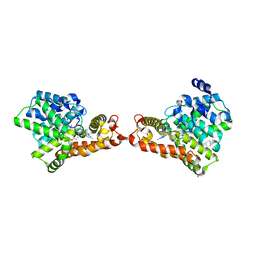 | | Crystal structure of human phosphodiesterase 2A in complex with 6-methyl-N-(1-(4-(trifluoromethoxy)phenyl)propyl)pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Descriptor: | 6-methyl-N-[(1R)-1-[4-(trifluoromethyloxy)phenyl]propyl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Oki, H, Kondo, M, Snell, G, Lane, W. | | Deposit date: | 2017-05-08 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of an Orally Bioavailable, Brain-Penetrating, in Vivo Active Phosphodiesterase 2A Inhibitor Lead Series for the Treatment of Cognitive Disorders.
J. Med. Chem., 60, 2017
|
|
8G2Y
 
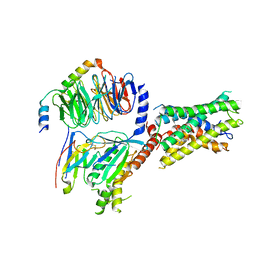 | | Cryo-EM structure of ADGRF1 coupled to miniGs/q | | Descriptor: | Adhesion G-protein-coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jones, D, Rawson, S, Blacklow, S. | | Deposit date: | 2023-02-06 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Tethered agonist activated ADGRF1 structure and signalling analysis reveal basis for G protein coupling.
Nat Commun, 14, 2023
|
|
3KJM
 
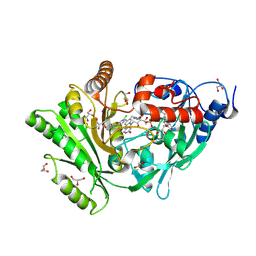 | |
