2C9D
 
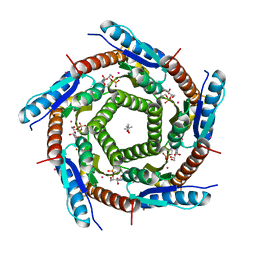 | | Lumazine Synthase from Mycobacterium tuberculosis Bound to 3-(1,3,7- TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL)HEXANE 1-PHOSPHATE | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(1,3,7-TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL ) HEXANE 1-PHOSPHATE, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, ... | | Authors: | Morgunova, E, Illarionov, B, Jin, G, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Thermodynamic Insights Into the Binding Mode of Five Novel Inhibitors of Lumazine Synthase from Mycobacterium Tuberculosis.
FEBS J., 273, 2006
|
|
2C94
 
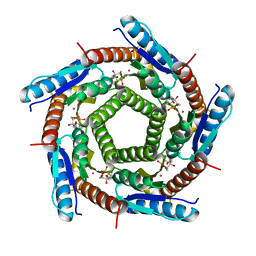 | | LUMAZINE SYNTHASE FROM MYCOBACTERIUM TUBERCULOSIS BOUND TO 3-(1,3,7- TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL) 1,1 difluoropentane-1- PHOSPHATE | | Descriptor: | 3-(1,3,7-TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL) 1,1 DIFLUOROPENTANE-1-PHOSPHATE, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, POTASSIUM ION | | Authors: | Morgunova, E, Illarionov, B, Jin, G, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Thermodynamic Insights Into the Binding Mode of Five Novel Inhibitors of Lumazine Synthase from Mycobacterium Tuberculosis.
FEBS J., 273, 2006
|
|
2C92
 
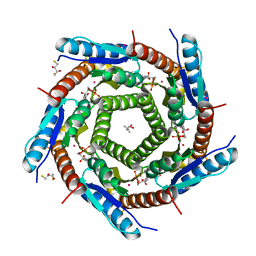 | | LUMAZINE SYNTHASE FROM MYCOBACTERIUM TUBERCULOSIS BOUND TO 3-(1,3,7- TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL) PENTANE 1 PHOSPHATE | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(1,3,7-TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL) 1-PHOSPHATE, ... | | Authors: | Morgunova, E, Illarionov, B, Jin, G, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Thermodynamic Insights Into the Binding Mode of Five Novel Inhibitors of Lumazine Synthase from Mycobacterium Tuberculosis.
FEBS J., 273, 2006
|
|
2C97
 
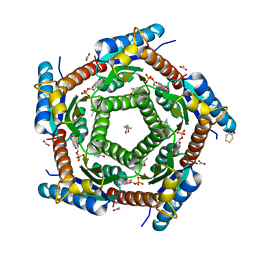 | | LUMAZINE SYNTHASE FROM MYCOBACTERIUM TUBERCULOSIS BOUND TO 4-(6- chloro-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)butyl phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(6-CHLORO-2,4-DIOXO-1,2,3,4-TETRAHYDROPYRIMIDIN-5-YL) BUTYL PHOSPHATE, 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, ... | | Authors: | Morgunova, E, Illarionov, B, Jin, G, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Thermodynamic Insights Into the Binding Mode of Five Novel Inhibitors of Lumazine Synthase from Mycobacterium Tuberculosis.
FEBS J., 273, 2006
|
|
4NLE
 
 | |
2C9B
 
 | | Lumazine Synthase from Mycobacterium tuberculosus Bound to 3-(1,3,7- TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(1,3,7-TRIHYDRO-9-D-RIBITYL-2,6,8-PURINETRIONE-7-YL), 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE, ... | | Authors: | Morgunova, E, Illarionov, B, Jin, G, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2005-12-09 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural and Thermodynamic Insights Into the Binding Mode of Five Novel Inhibitors of Lumazine Synthase from Mycobacterium Tuberculosis.
FEBS J., 273, 2006
|
|
5GQO
 
 | |
3RUX
 
 | |
3ZHR
 
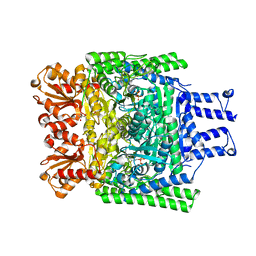 | | Crystal structure of the H747A mutant of the SucA domain of Mycobacterium smegmatis KGD showing the active site lid closed | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
2YIC
 
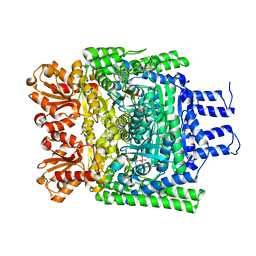 | | Crystal structure of the SucA domain of Mycobacterium smegmatis alpha- ketoglutarate decarboxylase (triclinic form) | | Descriptor: | 2-OXOGLUTARATE DECARBOXYLASE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
1TW2
 
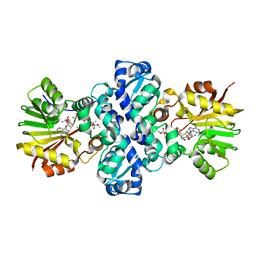 | | Crystal structure of Carminomycin-4-O-methyltransferase (DnrK) in complex with S-adenosyl-L-homocystein (SAH) and 4-methoxy-e-rhodomycin T (M-ET) | | Descriptor: | Carminomycin 4-O-methyltransferase, METHYL (4R)-2-ETHYL-2,5,12-TRIHYDROXY-7-METHOXY-6,11-DIOXO-4-{[2,3,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-RIBO-HEXOPYRANOSYL]OXY}-1H,2H,3H,4H,6H,11H-TETRACENE-1-CARBOXYLATE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Jansson, A, Koskiniemi, H, Mantsala, P, Niemi, J, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-30 | | Release date: | 2004-09-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a ternary complex of DnrK, a methyltransferase in daunorubicin biosynthesis, with bound products
J.Biol.Chem., 279, 2004
|
|
3E25
 
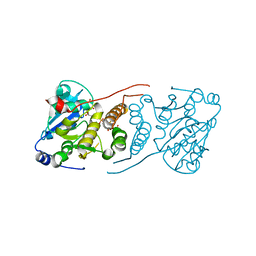 | | Crystal structure of M. tuberculosis glucosyl-3-phosphoglycerate synthase | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, MAGNESIUM ION, Putative uncharacterized protein, ... | | Authors: | Pereira, P.J.B, Empadinhas, N, Costa, M.S, Macedo-Ribeiro, S. | | Deposit date: | 2008-08-05 | | Release date: | 2008-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mycobacterium tuberculosis glucosyl-3-phosphoglycerate synthase: structure of a key enzyme in methylglucose lipopolysaccharide biosynthesis
Plos One, 3, 2008
|
|
2EV1
 
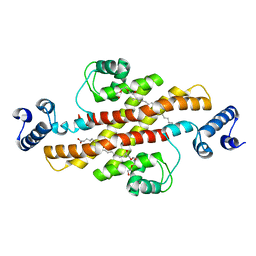 | | Structure of Rv1264N, the regulatory domain of the mycobacterial adenylyl cylcase Rv1264, at pH 6.0 | | Descriptor: | Hypothetical protein Rv1264/MT1302, OLEIC ACID, PENTAETHYLENE GLYCOL | | Authors: | Findeisen, F, Tews, I, Sinning, I. | | Deposit date: | 2005-10-30 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the regulatory domain of the adenylyl cyclase Rv1264 from Mycobacterium tuberculosis with bound oleic acid
J.Mol.Biol., 369, 2007
|
|
2EV3
 
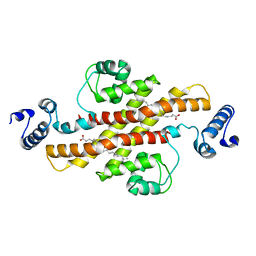 | | Structure of Rv1264N, the regulatory domain of the mycobacterial adenylyl cylcase Rv1264, at pH 5.3 | | Descriptor: | Hypothetical protein Rv1264/MT1302, OLEIC ACID | | Authors: | Findeisen, F, Tews, I, Sinning, I. | | Deposit date: | 2005-10-30 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The structure of the regulatory domain of the adenylyl cyclase Rv1264 from Mycobacterium tuberculosis with bound oleic acid
J.Mol.Biol., 369, 2007
|
|
1WQG
 
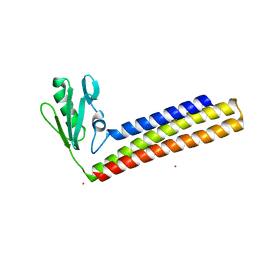 | | Crystal structure of ribosome recycling factor from Mycobacterium Tuberculosis | | Descriptor: | CADMIUM ION, Ribosome recycling factor | | Authors: | Saikrishnan, K, Kalapala, S.K, Varshney, U, Vijayan, M. | | Deposit date: | 2004-09-29 | | Release date: | 2005-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray structural studies of Mycobacterium tuberculosis RRF and a comparative study of RRFs of known structure. Molecular plasticity and biological implications
J.Mol.Biol., 345, 2005
|
|
4OIZ
 
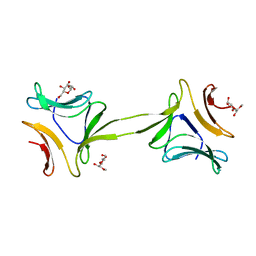 | | Structure, interactions and evolutionary implications of a domain-swapped lectin dimer from Mycobacterium smegmatis | | Descriptor: | LysM domain protein, methyl alpha-D-mannopyranoside | | Authors: | Patra, D, Mishra, P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-01-20 | | Release date: | 2014-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure, interactions and evolutionary implications of a domain-swapped lectin dimer from Mycobacterium smegmatis.
Glycobiology, 24, 2014
|
|
4OIT
 
 | | Structure, interactions and evolutionary implications of a domain-swapped lectin dimer from Mycobacterium smegmatis | | Descriptor: | LysM domain protein, alpha-D-mannopyranose, beta-D-mannopyranose | | Authors: | Patra, D, Mishra, P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-01-20 | | Release date: | 2014-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure, interactions and evolutionary implications of a domain-swapped lectin dimer from Mycobacterium smegmatis.
Glycobiology, 24, 2014
|
|
2EV2
 
 | | Structure of Rv1264N, the regulatory domain of the mycobacterial adenylyl cylcase Rv1264, at pH 8.5 | | Descriptor: | Hypothetical protein Rv1264/MT1302, OLEIC ACID | | Authors: | Findeisen, F, Tews, I, Sinning, I. | | Deposit date: | 2005-10-30 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of the regulatory domain of the adenylyl cyclase Rv1264 from Mycobacterium tuberculosis with bound oleic acid
J.Mol.Biol., 369, 2007
|
|
1WQF
 
 | | Crystal structure of Ribosome recycling factor from Mycobacterium Tuberculosis | | Descriptor: | CADMIUM ION, Ribosome recycling factor | | Authors: | Saikrishnan, K, Kalapala, S.K, Varshney, U, Vijayan, M. | | Deposit date: | 2004-09-28 | | Release date: | 2005-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | X-ray structural studies of Mycobacterium tuberculosis RRF and a comparative study of RRFs of known structure. Molecular plasticity and biological implications
J.Mol.Biol., 345, 2005
|
|
1WQH
 
 | | Crystal structure of ribosome recycling factor from Mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, Ribosome recycling factor | | Authors: | Saikrishnan, K, Kalapala, S.K, Varshney, U, Vijayan, M. | | Deposit date: | 2004-09-29 | | Release date: | 2005-02-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structural studies of Mycobacterium Tuberculosis RRF and a comparative study of RRFS of known structure. Molecular plasticity and biological implications
J.Mol.Biol., 345, 2005
|
|
2ZJ0
 
 | | Crystal structure of Mycobacterium tuberculosis S-Adenosyl-L-homocysteine hydrolase in ternary complex with NAD and 2-fluoroadenosine | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Reddy, M.C.M, Gokulan, K, Shetty, N.D, Owen, J.L, Ioerger, T.R, Sacchettini, J.C. | | Deposit date: | 2008-02-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis S-adenosyl-L-homocysteine hydrolase in ternary complex with substrate and inhibitors.
Protein Sci., 17, 2008
|
|
8I61
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Barbituric acid and Citric acid, Form I | | Descriptor: | 1,2-ETHANEDIOL, BARBITURIC ACID, CITRIC ACID, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I69
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 5-Fluoroorotic acid and Citric acid, Form I | | Descriptor: | 1,2-ETHANEDIOL, 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CITRIC ACID, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I67
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 2,4-Thiazolidinedione, Form I | | Descriptor: | 1,2-ETHANEDIOL, 1,3-thiazolidine-2,4-dione, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
8I6B
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with 5-Hydroxy-2,4(1H,3H)-pyrimidinedione, Form I | | Descriptor: | 1,2-ETHANEDIOL, 5-oxidanyl-1~{H}-pyrimidine-2,4-dione, CHLORIDE ION, ... | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
