5VLD
 
 | |
3CBE
 
 | |
5WZQ
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - quadruple mutant | | Descriptor: | Alpha-N-acetylgalactosaminidase, GLYCEROL, ZINC ION | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
8B26
 
 | | Dihydroprecondylocarpine acetate synthase 2 from Tabernanthe iboga | | Descriptor: | Dihydroprecondylocarpine acetate synthase 2, ZINC ION | | Authors: | Langley, C, Basquin, J, Caputi, L, O'Connor, S.E. | | Deposit date: | 2022-09-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Expansion of the Catalytic Repertoire of Alcohol Dehydrogenases in Plant Metabolism.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8AD7
 
 | |
8AE3
 
 | | Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgM C2-domain from mouse, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-07-12 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
8DOC
 
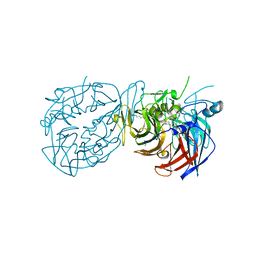 | | Crystal structure of RPE65 in complex with compound 16e and palmitate | | Descriptor: | (1R)-1-[3-(cyclohexylmethoxy)phenyl]-3-(methylamino)propan-1-ol, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, FE (II) ION, ... | | Authors: | Bassetto, M, Kiser, P.D. | | Deposit date: | 2022-07-12 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tuning the Metabolic Stability of Visual Cycle Modulators through Modification of an RPE65 Recognition Motif.
J.Med.Chem., 66, 2023
|
|
3NE7
 
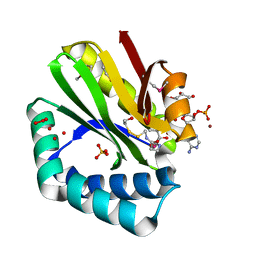 | | Crystal structure of paia n-acetyltransferase from thermoplasma acidophilum in complex with coenzyme a | | Descriptor: | ACETYLTRANSFERASE, BETA-MERCAPTOETHANOL, COENZYME A, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
6WBT
 
 | | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-alpha-D-glucopyranose, MANGANESE (II) ION, ... | | Authors: | Wu, R, Kim, Y, Endres, M, Joachimiak, J. | | Deposit date: | 2020-03-27 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | 2.52 Angstrom Resolution Crystal Structure of 6-phospho-alpha-glucosidase from Gut Microorganisms in Complex with NAD and Glucose-6-phosphate
To Be Published
|
|
1TRE
 
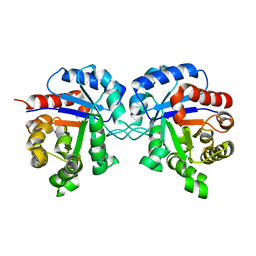 | |
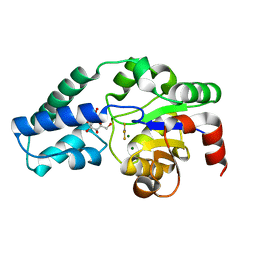
5O6R
 
 | |
5AKF
 
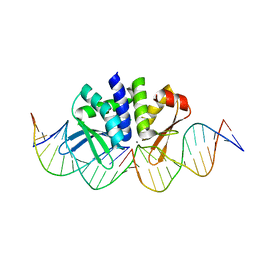 | | THE CRYSTAL STRUCTURE OF I-DMOI Q42AK120M IN COMPLEX WITH ITS TARGET DNA NICKED IN THE CODING STRAND A AND IN THE PRESENCE OF 2MM MN | | Descriptor: | 25MER, 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP)-3', 5'-D(*GP*TP*TP*CP*CP*GP*GP*CP*GP*CP*GP)-3', ... | | Authors: | Molina, R, Marcaida, M.J, Redondo, P, Marenchino, M, D'Abramo, M, Montoya, G, Prieto, J. | | Deposit date: | 2015-03-03 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Engineering a Nickase on the Homing Endonuclease I-Dmoi Scaffold.
J.Biol.Chem., 290, 2015
|
|
8JU4
 
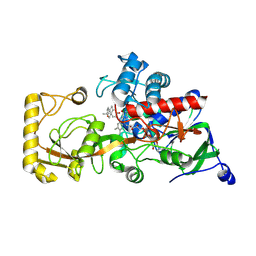 | | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione | | Descriptor: | 3-ketosteroid dehydrogenase, ANDROSTA-1,4-DIENE-3,17-DIONE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hu, Y.L, Li, X, Cheng, X.Y, Song, S.K, Su, Z.D. | | Deposit date: | 2023-06-24 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of 3-ketosteroid delta1-dehydrogenase from Rhodococcus erythropolis SQ1 in complex with 1,4-androstadiene-3,17- dione
To Be Published
|
|
8ADZ
 
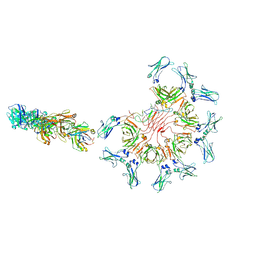 | | Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgM C2-domain from mouse, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-07-12 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
8AE0
 
 | | Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgM C2-domain from mouse, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-07-12 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
8R06
 
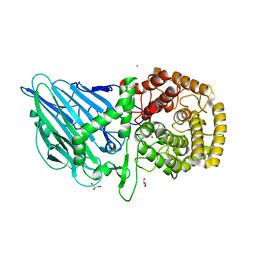 | | CRYSTAL STRUCTURE OF THERMOANAEROBACTERIUM XYLANOLYTICUM GH116 BETA-GLUCOSIDASE WITH A COVALENTLY BOUND CYCLOPHELLITOL AZIRIDINE | | Descriptor: | (1~{R},2~{S},3~{S},4~{S},5~{R},6~{R})-5-azanyl-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Offen, W.A, Davies, G.J, Breen, I.Z. | | Deposit date: | 2023-10-30 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CRYSTAL STRUCTURE OF THERMOANAEROBACTERIUM XYLOLYTICUM GH116 BETA-GLUCOSIDASE WITH A COVALENTLY BOUND CYCLOPHELLITOL AZIRIDINE
JACS, 40, 2017
|
|
8ADY
 
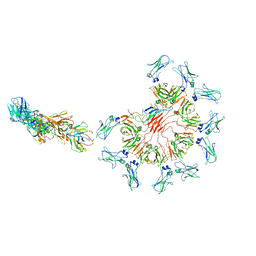 | | Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgM C2-domain from mouse, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-07-12 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
6W49
 
 | | Trypanosoma cruzi Malic Enzyme in complex with inhibitor (MEC010) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRIC ACID, Malic enzyme, ... | | Authors: | Mercaldi, G.F, Fagundes, M, Faria, J.N, Cordeiro, A.T. | | Deposit date: | 2020-03-10 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Trypanosoma cruzi Malic Enzyme Is the Target for Sulfonamide Hits from the GSK Chagas Box.
Acs Infect Dis., 7, 2021
|
|
8KDE
 
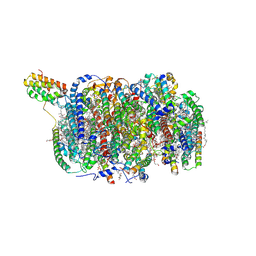 | | Cryo-EM structure of an intermediate-state complex during the process of photosystem II repair | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, A, Wang, Y, Liu, Z. | | Deposit date: | 2023-08-09 | | Release date: | 2024-06-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for an early stage of the photosystem II repair cycle in Chlamydomonas reinhardtii.
Nat Commun, 15, 2024
|
|
6W57
 
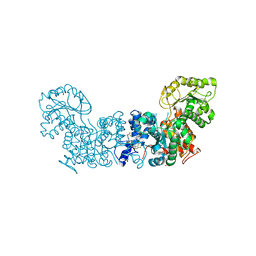 | | Trypanosoma cruzi Malic Enzyme in complex with inhibitor (MEC069) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Malic enzyme, ~{N}-[5-[[4-[bis(fluoranyl)methoxy]phenyl]sulfamoyl]-2-chloranyl-phenyl]-3,5-bis(fluoranyl)benzamide | | Authors: | Mercaldi, G.F, Fagundes, M, Faria, J.N, Cordeiro, A.T. | | Deposit date: | 2020-03-12 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Trypanosoma cruzi Malic Enzyme Is the Target for Sulfonamide Hits from the GSK Chagas Box.
Acs Infect Dis., 7, 2021
|
|
4ZX3
 
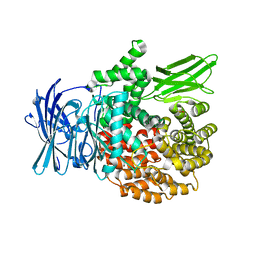 | |
5WZR
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - Gal-NHAc-DNJ complex | | Descriptor: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, N-[(3S,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]acetamide, ... | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
6W53
 
 | | Trypanosoma cruzi Malic Enzyme in complex with inhibitor (MEC070) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Malic enzyme, N-(2-chloro-5-{[4-(trifluoromethoxy)phenyl]sulfamoyl}phenyl)-3,5-difluorobenzamide | | Authors: | Mercaldi, G.F, Fagundes, M, Faria, J.N, Cordeiro, A.T. | | Deposit date: | 2020-03-12 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trypanosoma cruzi Malic Enzyme Is the Target for Sulfonamide Hits from the GSK Chagas Box.
Acs Infect Dis., 7, 2021
|
|
1AKC
 
 | |
8AE2
 
 | | Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgM C2-domain from mouse, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-07-12 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
