3EUM
 
 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6066 | | Descriptor: | 3,6-Bis[3-(azepan-1-yl)propionamido]acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3EUI
 
 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6042 in a large unit cell | | Descriptor: | 3,6-Bis{3-[(2R)-(2-ethylpiperidino)]propionamido}acridine, 3-[(2R)-2-ethylpiperidin-1-yl]-N-[6-({3-[(2S)-2-ethylpiperidin-1-yl]propanoyl}amino)acridin-3-yl]propanamide, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', ... | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-10 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3ERU
 
 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6045 | | Descriptor: | 3,6-Bis{3-[(2R)-2-methylpiperidino)]propionamido}acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-03 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3ES0
 
 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6048 | | Descriptor: | 3,6-Bis[3-(4-methylpiperidino)propionamido]acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-03 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
4O9D
 
 | | Structure of Dos1 propeller | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Rik1-associated factor 1 | | Authors: | Kuscu, C, Schalch, T, Joshua-Tor, L. | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRL4-like Clr4 complex in Schizosaccharomyces pombe depends on an exposed surface of Dos1 for heterochromatin silencing.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2REU
 
 | | Crystal Structure of the C-terminal of Sau3AI fragment | | Descriptor: | MAGNESIUM ION, Type II restriction enzyme Sau3AI | | Authors: | Hu, X, Yu, F, Xu, C, He, J. | | Deposit date: | 2007-09-27 | | Release date: | 2008-09-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and function of C-terminal Sau3AI domain
Biochim.Biophys.Acta, 1794, 2009
|
|
6X1B
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with the Product Nucleotide GpU. | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-R(*GP*U)-3'), PHOSPHATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-18 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6Q44
 
 | | Est3 telomerase subunit in the yeast Hansenula polymorpha | | Descriptor: | Uncharacterized protein | | Authors: | Mantsyzov, A.B, Mariasina, S.S, Petrova, O.A, Efimov, S.V, Dontsova, O.A, Polshakov, V.I. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-25 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Insights into the structure and function of Est3 from the Hansenula polymorpha telomerase.
Sci Rep, 10, 2020
|
|
6QXH
 
 | |
6QXG
 
 | |
1TP4
 
 | |
6R2E
 
 | | Crystal structure of the human thymidylate synthase (hTS) interface variant Q62R | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Pozzi, C, Mangani, M. | | Deposit date: | 2019-03-16 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Evidence of Destabilization of the Human Thymidylate Synthase (hTS) Dimeric Structure Induced by the Interface Mutation Q62R.
Biomolecules, 9, 2019
|
|
5XBL
 
 | | Structure of nuclease in complex with associated protein | | Descriptor: | Associated protein, CRISPR-associated endonuclease Cas9/Csn1, RNA (98-MER) | | Authors: | Dong, D, Guo, M, Wang, S, Zhu, Y, Huang, Z. | | Deposit date: | 2017-03-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein
Nature, 546, 2017
|
|
8PSN
 
 | |
8PSO
 
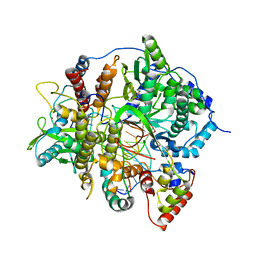 | | Tilapia Lake Virus polymerase in vRNA initiation state (core only) | | Descriptor: | 5' vRNA end - vRNA loop (40-mer), DNA (5'-D(*(CTP))-3'), MAGNESIUM ION, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2023-07-13 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural and functional analysis of the minimal orthomyxovirus-like polymerase of Tilapia Lake Virus from the highly diverged Amnoonviridae family.
Nat Commun, 14, 2023
|
|
8PT6
 
 | |
8FR5
 
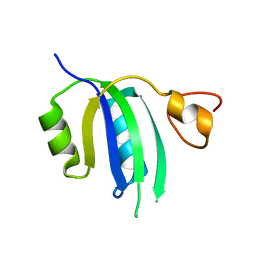 | | Crystal structure of the Human Smacovirus 1 Rep domain | | Descriptor: | MANGANESE (II) ION, Rep, SODIUM ION | | Authors: | Limon, L.K, Shi, K, Dao, A, Rugloski, J, Tompkins, K.J, Aihara, H, Gordon, W.R, Evans IIII, R.L. | | Deposit date: | 2023-01-06 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | The crystal structure of the human smacovirus 1 Rep domain.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
4UKD
 
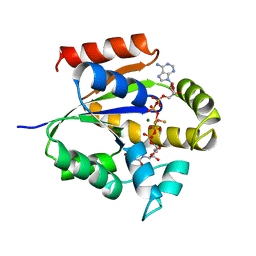 | | UMP/CMP KINASE FROM SLIME MOLD COMPLEXED WITH ADP, UDP, BERYLLIUM FLUORIDE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM DIFLUORIDE, MAGNESIUM ION, ... | | Authors: | Schlichting, I, Reinstein, J. | | Deposit date: | 1997-05-20 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of active conformations of UMP kinase from Dictyostelium discoideum suggest phosphoryl transfer is associative.
Biochemistry, 36, 1997
|
|
4V7N
 
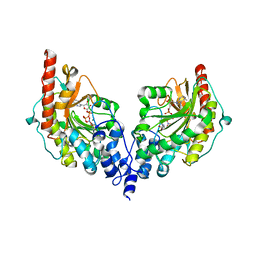 | | Glycocyamine kinase, beta-beta homodimer from marine worm Namalycastis sp., with transition state analog Mg(II)-ADP-NO3-glycocyamine. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANIDINO ACETATE, Glycocyamine kinase beta chain, ... | | Authors: | Lim, K, Pullalarevu, S, Herzberg, O. | | Deposit date: | 2009-12-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the mechanism and substrate specificity of glycocyamine kinase, a phosphagen kinase family member.
Biochemistry, 49, 2010
|
|
1U6N
 
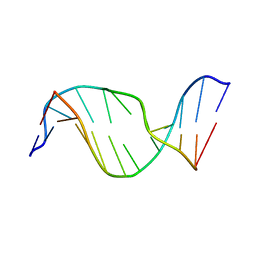 | | Solution Structure of an Oligodeoxynucleotide Containing a Butadiene Derived N1 b-Hydroxyalkyl Adduct on Deoxyinosine in the Human N-ras Codon 61 Sequence | | Descriptor: | 5'-D(*CP*GP*GP*AP*CP*(2BD)P*AP*GP*AP*AP*G)-3', 5'-D(*CP*TP*TP*CP*TP*TP*GP*TP*CP*CP*G)-3' | | Authors: | Scholdberg, T.A, Merritt, W.K, Dean, S.M, Kowalcyzk, A, Harris, T.M, Harris, C.M, Lloyd, R.S, Stone, M.P. | | Deposit date: | 2004-07-30 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an Oligodeoxynucleotide Containing a Butadiene Oxide-Derived N1 Beta-Hydroxyalkyl Deoxyinosine Adduct in the Human N-ras Codon 61 Sequence.
Biochemistry, 44, 2005
|
|
1Z96
 
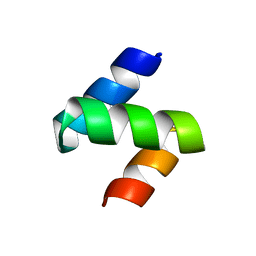 | | Crystal structure of the Mud1 UBA domain | | Descriptor: | UBA-domain protein mud1 | | Authors: | Trempe, J.-F, Brown, N.R, Lowe, E.D, Noble, M.E.M, Gordon, C, Campbell, I.D, Johnson, L.N, Endicott, J.A. | | Deposit date: | 2005-03-31 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Lys48-linked polyubiquitin chain recognition by the Mud1 UBA domain
Embo J., 24, 2005
|
|
8G3D
 
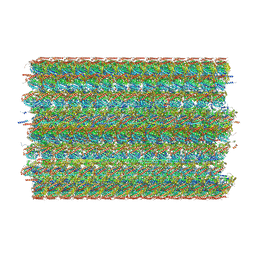 | | 48-nm doublet microtubule from Tetrahymena thermophila strain K40R | | Descriptor: | B2B3_fMIP, B5B6_fMIP, CFAM166A, ... | | Authors: | Black, C.S, Kubo, S, Yang, S.K, Bui, K.H. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Native doublet microtubules from Tetrahymena thermophila reveal the importance of outer junction proteins.
Nat Commun, 14, 2023
|
|
8G2Z
 
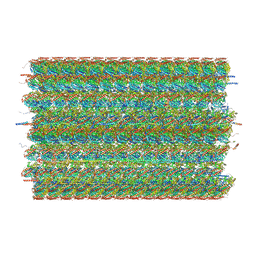 | | 48-nm doublet microtubule from Tetrahymena thermophila strain CU428 | | Descriptor: | B2B3_fMIP, B5B6_fMIP, CFAM166A, ... | | Authors: | Black, C.S, Kubo, S, Yang, S.K, Bui, K.H. | | Deposit date: | 2023-02-06 | | Release date: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Native doublet microtubules from Tetrahymena thermophila reveal the importance of outer junction proteins.
Nat Commun, 14, 2023
|
|
6XRQ
 
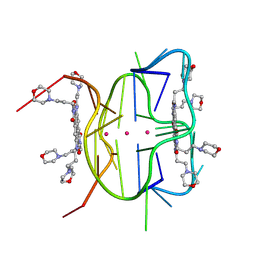 | | Structural descriptions of ligand interactions to DNA and RNA quadruplexes folded from the non-coding region of Pseudorabies virus | | Descriptor: | 2,7-bis[3-(morpholin-4-yl)propyl]-4,9-bis{[3-(morpholin-4-yl)propyl]amino}benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetrone, POTASSIUM ION, RNA (5' GP*GP*CP*UP*CP*GP*GP*CP*GP*GP*CP*GP*GP*A-3') | | Authors: | Zhang, Y.S, Parkinson, G.N, Wei, D.G. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural descriptions of ligand interactions to RNA quadruplexes folded from the non-coding region of Pseudorabies virus.
Biochimie, 2024
|
|
8K3Y
 
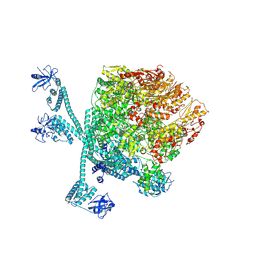 | | The "5+1" heteromeric structure of Lon protease consisting of a spiral pentamer with Y224S mutation and an N-terminal-truncated monomeric E613K mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Li, S, Hsieh, K.Y, Kuo, C.I, Zhang, K, Chang, C.I. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A 5+1 assemble-to-activate mechanism of the Lon proteolytic machine.
Nat Commun, 14, 2023
|
|
