6VLG
 
 | | Crystal structure of mouse alpha 1,6-fucosyltransferase, FUT8 bound to GDP | | Descriptor: | Alpha-(1,6)-fucosyltransferase, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Jarva, M.A, Dramicanin, M, Lingford, J.P, Mao, R, John, A, Goddard-Borger, E.D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of substrate recognition and catalysis by fucosyltransferase 8.
J.Biol.Chem., 295, 2020
|
|
6VXR
 
 | | Structure of Maternal embryonic leucine zipper kinase bound to LDSM276 | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-7-methyl-5,8-di(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, 1,2-ETHANEDIOL, Maternal embryonic leucine zipper kinase | | Authors: | Counago, R.M, Takarada, J.E, dos Reis, C.V, Gama, F.H, Azevedo, H, Mascarello, A, Guimaraes, C.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-24 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Maternal embryonic leucine zipper kinase bound to LDSM276
To Be Published
|
|
6VYC
 
 | | Crystal structure of WD-repeat domain of human WDR91 | | Descriptor: | UNKNOWN ATOM OR ION, WD repeat-containing protein 91 | | Authors: | Halabelian, L, Hutchinson, A, Li, Y, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of a First-in-Class Small-Molecule Ligand for WDR91 Using DNA-Encoded Chemical Library Selection Followed by Machine Learning.
J.Med.Chem., 66, 2023
|
|
7KQK
 
 | |
6W4Q
 
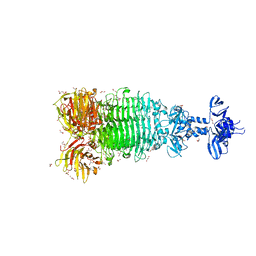 | | Crystal structure of full-length tailspike protein 2 (TSP2, ORF211) ) from Escherichia coli O157:H7 bacteriophage CAB120 | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Greenfield, J, Herzberg, O. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of bacteriophage CBA120 ORF211 (TSP2), the determinant of phage specificity towards E. coli O157:H7.
Sci Rep, 10, 2020
|
|
7MEQ
 
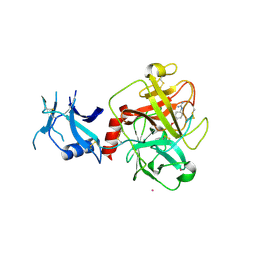 | | Crystal structure of human TMPRSS2 in complex with Nafamostat | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-carbamimidamidobenzoic acid, Transmembrane protease serine 2, ... | | Authors: | Fraser, B, Beldar, S, Hutchinson, A, Li, Y, Seitova, A, Edwards, A.M, Benard, F, Arrowsmith, C.H, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-04-07 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and activity of human TMPRSS2 protease implicated in SARS-CoV-2 activation.
Nat.Chem.Biol., 18, 2022
|
|
7NKW
 
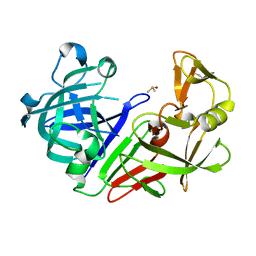 | | Endothiapepsin structure obtained at 298K after a soaking with fragment JFD03909 from a dataset collected with JUNGFRAU detector | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Engilberge, S, Huang, C.-Y, Leonarski, F, Wojdyla, J.A, Marsh, M, Olieric, V, Wang, M. | | Deposit date: | 2021-02-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Endothiapepsin structure obtained at 298K after a soaking with fragment JFD03909 from a dataset collected with JUNGFRAU detector
To Be Published
|
|
7N2X
 
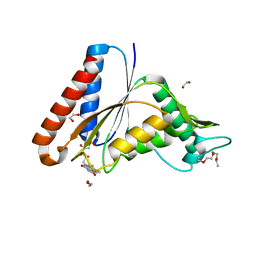 | | The crystal structure of an FMN-dependent NADH:quinone oxidoreductase, AzoR from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-AMINO-ACRYLIC ACID, ... | | Authors: | Arcinas, A.J, Fedorov, E, Kelly, L, Almo, S.C, Ghosh, A. | | Deposit date: | 2021-05-30 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Uncovering a novel mechanism of enzyme activation in multimeric azoreductases
To Be Published
|
|
6DPX
 
 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | Descriptor: | (3-{[(3-chloro-2-hydroxyphenyl)sulfonyl]amino}phenyl)acetic acid, Beta-lactamase | | Authors: | Singh, I. | | Deposit date: | 2018-06-09 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ultra-large library docking for discovering new chemotypes.
Nature, 566, 2019
|
|
6QM0
 
 | | Cathepsin-K in complex with amino-oxaazabicyclo[3.3.0]octanyl containing inhibitor | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Cathepsin K, ~{N}-[(2~{S})-1-[(3~{R},3~{a}~{R},6~{R},6~{a}~{R})-6-azanyl-3-oxidanyl-2,3,3~{a},5,6,6~{a}-hexahydrofuro[3,2-b]pyrrol-4-yl]-4-methyl-1-oxidanylidene-pentan-2-yl]-3-fluoranyl-4-[2-(4-methylpiperazin-1-yl)-1,3-thiazol-4-yl]benzamide | | Authors: | Derbyshire, D.J. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Successful development of 3-oxohexahydrofuropyrrole amino acid amides as inhibitors of Cathepsin-K.
To Be Published
|
|
6DXA
 
 | |
7AM5
 
 | |
6QLX
 
 | | Cathepsin-K in complex with fluoro-oxa-azabicyclo[3.3.0]octanyl containing inhibitor | | Descriptor: | Cathepsin K, ~{N}-[(2~{S})-1-[(3~{R},3~{a}~{R},6~{S},6~{a}~{S})-6-fluoranyl-3-oxidanyl-2,3,3~{a},5,6,6~{a}-hexahydrofuro[3,2-b]pyrrol-4-yl]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-thiophen-2-yl-benzamide | | Authors: | Derbyshire, D.J. | | Deposit date: | 2019-02-01 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Successful development of 3-oxohexahydrofuropyrrole amino acid amides as inhibitors of Cathepsin-K.
To Be Published
|
|
7AVZ
 
 | | MerTK kinase domain in complex with a bisaminopyrimidine inhibitor | | Descriptor: | (R)-N2-(4-(cyclopropylmethoxy)-3,5-difluorophenyl)-5-(3-methylpiperazin-1-yl)-N4-(tetrahydro-2H-pyran-4-yl)pyrimidine-2,4-diamine, Tyrosine-protein kinase Mer | | Authors: | Pflug, A, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Preston, M, Rawlins, P, Rivers, E, Schimpl, M, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
7AW2
 
 | | MerTK kinase domain with type 1.5 inhibitor from a DNA-encoded library | | Descriptor: | 5-(2'-chloro-[1,1'-biphenyl]-4-yl)-N-(imidazo[1,2-a]pyridin-6-ylmethyl)-N-methyl-1,3,4-oxadiazol-2-amine, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
6DXF
 
 | |
7AW1
 
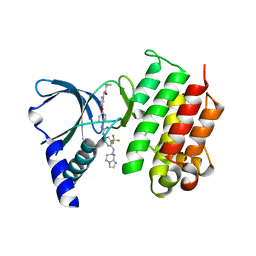 | | MerTK kinase domain in complex with a type 2 inhibitor | | Descriptor: | N-(6-(4-(3-(4-((5,6-dihydroimidazo[1,2-a]pyrazin-7(8H)-yl)methyl)-3-(trifluoromethyl)phenyl)ureido)phenoxy)pyrimidin-4-yl)cyclopropanecarboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
6QXL
 
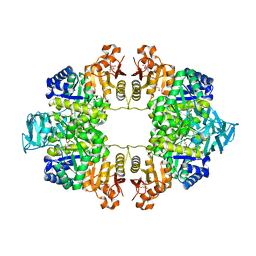 | | Crystal Structure of Pyruvate Kinase II (PykA) from Pseudomonas aeruginosa in complex with sodium malonate, magnesium and glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Abdelhamid, Y, Brear, P, Welch, M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Evolutionary plasticity in the allosteric regulator-binding site of pyruvate kinase isoform PykA fromPseudomonas aeruginosa.
J.Biol.Chem., 294, 2019
|
|
6QZJ
 
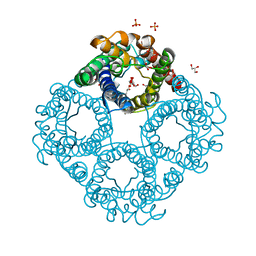 | | Crystal structure of human Aquaporin 7 at 2.2 A resolution | | Descriptor: | Aquaporin-7, GLYCEROL, PHOSPHATE ION | | Authors: | de Mare, S.W.-H, Venskutonyte, R, Eltschkner, S, Lindkvist-Petersson, K. | | Deposit date: | 2019-03-11 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Glycerol Efflux and Selectivity of Human Aquaporin 7.
Structure, 28, 2020
|
|
6D5A
 
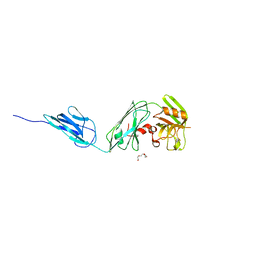 | |
6DNT
 
 | | UDP-N-acetylglucosamine 4-epimerase from Methanobrevibacter ruminantium M1 in complex with UDP-N-acetylmuramic acid | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase, ... | | Authors: | Carbone, V, Schofield, L.R, Sang, C, Sutherland-Smith, A.J, Ronimus, R.S. | | Deposit date: | 2018-06-07 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural determination of archaeal UDP-N-acetylglucosamine 4-epimerase from Methanobrevibacter ruminantium M1 in complex with the bacterial cell wall intermediate UDP-N-acetylmuramic acid.
Proteins, 86, 2018
|
|
6DPZ
 
 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | Descriptor: | (1R,2S)-2-{[(pyrrolidin-1-yl)sulfonyl]amino}cyclooctane-1-carboxylic acid, Beta-lactamase | | Authors: | Singh, I. | | Deposit date: | 2018-06-09 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ultra-large library docking for discovering new chemotypes.
Nature, 566, 2019
|
|
7AOU
 
 | | The Fk1 domain of FKBP51 in complex with (2'R,5'S,12'R)-12'-cyclohexyl-2'-[2-(3,4-dimethoxyphenyl)ethyl]-3',19'-dioxa-10',13',16'-triazaspiro[cyclopropane-1,15'- tricyclo[18.3.1.0-5,10]tetracosane]-1'(24'),20',22'-triene-4',11',14',17'-tetrone | | Descriptor: | (2'R,5'S,12'R)-12'-cyclohexyl-2'-[2-(3,4-dimethoxyphenyl)ethyl]-3',19'-dioxa-10',13',16'-triazaspiro[cyclopropane-1,15'- tricyclo[18.3.1.0-5,10]tetracosane]-1'(24'),20',22'-triene-4',11',14',17'-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, M.A, Meyners, C, Heymann, T, Merz, S, Purder, P, Bracher, A, Hausch, F. | | Deposit date: | 2020-10-15 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Macrocyclic FKBP51 Ligands Define a Transient Binding Mode with Enhanced Selectivity.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6QZI
 
 | | Crystal structure of human Aquaporin 7 at 1.9 A resolution | | Descriptor: | Aquaporin-7, GLYCEROL, PHOSPHATE ION | | Authors: | de Mare, S.W.-H, Venskutonyte, R, Eltschkner, S, Lindkvist-Petersson, K. | | Deposit date: | 2019-03-11 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Glycerol Efflux and Selectivity of Human Aquaporin 7.
Structure, 28, 2020
|
|
6D9C
 
 | |
