7ZWF
 
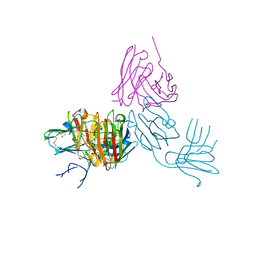 | | Pfs48/45 bound to scFv fragment of monoclonal antibody 32F3 | | Descriptor: | 32F3scFv, Gametocyte surface protein P45/48, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ko, K.T, Lennartz, F.L, Higgins, M.K. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.132 Å) | | Cite: | Structure of the malaria vaccine candidate Pfs48/45 and its recognition by transmission blocking antibodies.
Nat Commun, 13, 2022
|
|
8JYQ
 
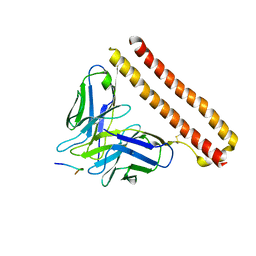 | |
8HGL
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-11 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
4Z95
 
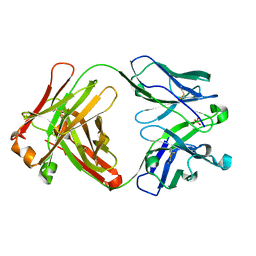 | |
4NKM
 
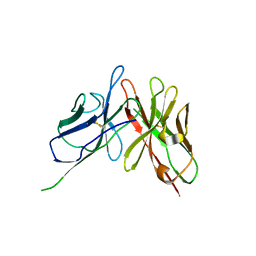 | | Crystal structure of engineered anti-EE scFv antibody fragment | | Descriptor: | Engineered scFv | | Authors: | Kalyoncu, S, Hyun, J, Pai, J.C, Johnson, J.L, Etzminger, K, Jain, A, Heaner Jr, D, Molares, I.A, Truskett, T.M, Maynard, J.A, Lieberman, R.L. | | Deposit date: | 2013-11-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Effects of protein engineering and rational mutagenesis on crystal lattice of single chain antibody fragments.
Proteins, 82, 2014
|
|
3V0W
 
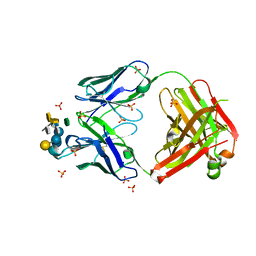 | | Crystal structure of Fab WN1 222-5 in complex with LPS | | Descriptor: | 2-amino-2-deoxy-alpha-D-glucopyranose-(1-2)-alpha-D-glucopyranose-(1-2)-alpha-D-glucopyranose-(1-3)-[alpha-D-galactopyranose-(1-6)]alpha-D-glucopyranose-(1-3)-[L-glycero-alpha-D-manno-heptopyranose-(1-7)]4-O-phosphono-L-glycero-alpha-D-manno-heptopyranose-(1-3)-4-O-phosphono-L-glycero-alpha-D-manno-heptopyranose-(1-5)-[3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)]3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, SULFATE ION, WN1 222-5 Fab (IgG2a) heavy chain, ... | | Authors: | Gomery, K, Evans, S.V. | | Deposit date: | 2011-12-08 | | Release date: | 2012-12-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Antibody WN1 222-5 mimics Toll-like receptor 4 binding in the recognition of LPS.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3V0V
 
 | | Fab WN1 222-5 unliganded | | Descriptor: | WN1 222-5 Fab (IgG2a) heavy chain, WN1 222-5 Fab (IgG2a) light chain | | Authors: | Gomery, K, Evans, S.V. | | Deposit date: | 2011-12-08 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Antibody WN1 222-5 mimics Toll-like receptor 4 binding in the recognition of LPS.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4M3J
 
 | | Structure of a single-domain camelid antibody fragment cAb-H7S specific of the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Camelid heavy-chain antibody variable fragment cAb-H7S, SULFATE ION | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
6Z3P
 
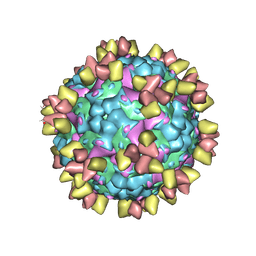 | | Structure of EV71 in complex with a protective antibody 38-3-11A Fab | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Zhou, D, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and functional analysis of protective antibodies targeting the threefold plateau of enterovirus 71.
Nat Commun, 11, 2020
|
|
5OCX
 
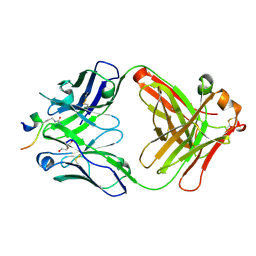 | | Crystal structure of ACPA E4 in complex with CII-C-13-CIT | | Descriptor: | 1,2-ETHANEDIOL, CII-C-13-CIT, Fab fragment anti-citrullinated protein antibody E4 - light chain, ... | | Authors: | Dobritzsch, D, Ge, C, Holmdahl, R. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies.
Arthritis Rheumatol, 71, 2019
|
|
4ZYK
 
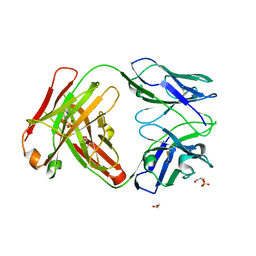 | | Crystal Structure of Quaternary-Specific RSV-Neutralizing Human Antibody AM14 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, AM14 Fab heavy chain, ... | | Authors: | Gilman, M.S.A, McLellan, J.S. | | Deposit date: | 2015-05-21 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein.
Plos Pathog., 11, 2015
|
|
6E0P
 
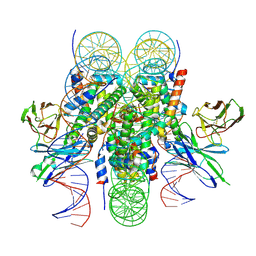 | |
6Z3Q
 
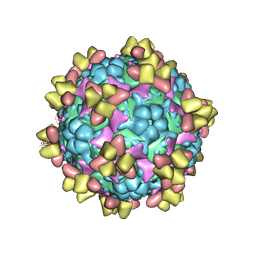 | | Structure of EV71 in complex with a protective antibody 38-1-10A Fab | | Descriptor: | Heavy chain, Light chain, SPHINGOSINE, ... | | Authors: | Zhou, D, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural and functional analysis of protective antibodies targeting the threefold plateau of enterovirus 71.
Nat Commun, 11, 2020
|
|
6BF4
 
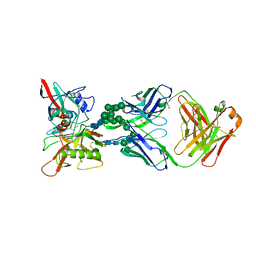 | |
6II9
 
 | | Crystal structure of H7 hemagglutinin from A/Anhui/1/2013 in complex with a human neutralizing antibody L3A-44 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of L3A-44 Fab, Hemagglutinin, ... | | Authors: | Jiang, H.H, Shi, Y, Qi, J, Gao, G.F. | | Deposit date: | 2018-10-03 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-function analysis of neutralizing antibodies to H7N9 influenza from naturally infected humans.
Nat Microbiol, 4, 2019
|
|
7R40
 
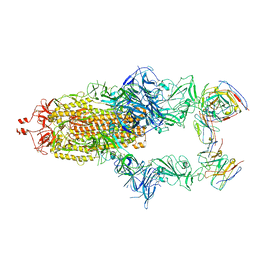 | | Structure of the SARS-CoV-2 spike glycoprotein in complex with the 87G7 antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 87G7 heavy chain variable region, ... | | Authors: | Hurdiss, D.L. | | Deposit date: | 2022-02-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | An ACE2-blocking antibody confers broad neutralization and protection against Omicron and other SARS-CoV-2 variants of concern.
Sci Immunol, 7, 2022
|
|
6CA7
 
 | |
3J91
 
 | | Cryo-electron microscopy of Enterovirus 71 (EV71) procapsid in complex with Fab fragments of neutralizing antibody 22A12 | | Descriptor: | VP0, VP1, VP3 | | Authors: | Shingler, K.L, Cifuente, J.O, Ashley, R.E, Makhov, A.M, Conway, J.F, Hafenstein, S. | | Deposit date: | 2014-11-24 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | The enterovirus 71 procapsid binds neutralizing antibodies and rescues virus infection in vitro.
J.Virol., 89, 2015
|
|
7K8N
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C102 | | Descriptor: | C102 Fab Heavy Chain, C102 Fab Light Chain, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8Q
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C121 | | Descriptor: | C121 Fab Heavy Chain, C121 Fab Light Chain, GLYCEROL | | Authors: | Abernathy, M.E, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
6XC3
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CC12.1 and CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.1 heavy chain, CC12.1 light chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
5FGC
 
 | |
6XC7
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with antibodies CC12.3 and CR3022 | | Descriptor: | CC12.3 heavy chain, CC12.3 light chain, CR3022 heavy chain, ... | | Authors: | Yuan, M, Liu, H, Wu, N.C, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.883 Å) | | Cite: | Structural basis of a shared antibody response to SARS-CoV-2.
Science, 369, 2020
|
|
7K8P
 
 | |
3J2Z
 
 | |
