5CDQ
 
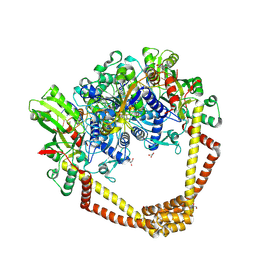 | | 2.95A structure of Moxifloxacin with S.aureus DNA gyrase and DNA | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA (5'-D(P*GP*AP*GP*CP*GP*TP*AP*T*GP*GP*CP*CP*AP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D, Srikannathasan, V, Chan, P.F. | | Deposit date: | 2015-07-04 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of DNA gyrase inhibition by antibacterial QPT-1, anticancer drug etoposide and moxifloxacin.
Nat Commun, 6, 2015
|
|
5COO
 
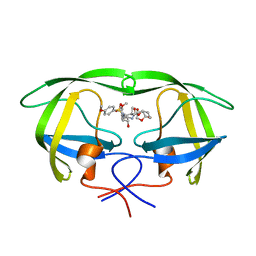 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-085 | | Descriptor: | (3R,3aS,4S,7aS)-3-hydroxyhexahydro-4H-furo[2,3-b]pyran-4-yl [(2S,3R)-3-hydroxy-1-(4-methoxyphenyl)-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}butan-2-yl]carbamate, HIV-1 protease | | Authors: | Yedidi, R.S, Hayashi, H, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2015-07-20 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | C-5-Modified Tetrahydropyrano-Tetrahydofuran-Derived Protease Inhibitors (PIs) Exert Potent Inhibition of the Replication of HIV-1 Variants Highly Resistant to Various PIs, including Darunavir.
J.Virol., 90, 2015
|
|
4IBM
 
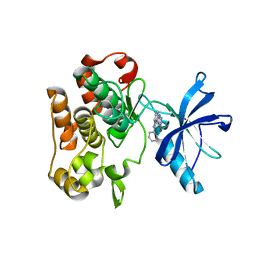 | | Crystal structure of insulin receptor kinase domain in complex with an inhibitor Irfin-1 | | Descriptor: | 5-(2-phenylpyrazolo[1,5-a]pyridin-3-yl)-3H-pyrazolo[3,4-c]pyridazin-3-one, Insulin receptor | | Authors: | Wu, J, Anastassiadis, T, Duong-Ly, K.C, Peterson, J.R. | | Deposit date: | 2012-12-08 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A highly selective dual insulin receptor (IR)/insulin-like growth factor 1 receptor (IGF-1R) inhibitor derived from an extracellular signal-regulated kinase (ERK) inhibitor.
J.Biol.Chem., 288, 2013
|
|
4QGI
 
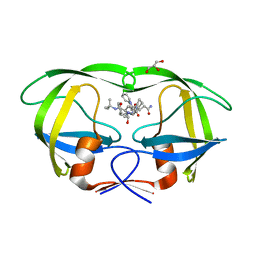 | | X-ray crystal structure of HIV-1 protease variant G48T/L89M in complex with Saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, GLYCEROL, Protease | | Authors: | Mahon, B.P, McKenna, R, Goldfarb, N. | | Deposit date: | 2014-05-22 | | Release date: | 2014-07-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Defective Hydrophobic Sliding Mechanism and Active Site Expansion in HIV-1 Protease Drug Resistant Variant Gly48Thr/Leu89Met: Mechanisms for the Loss of Saquinavir Binding Potency.
Biochemistry, 54, 2015
|
|
3I7C
 
 | |
6KZC
 
 | | crystal structure of TRKc in complex with 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methyl-N-(3-((4- methylpiperazin-1-yl)methyl)-5- (trifluoromethyl)phenyl)benzamide | | Descriptor: | 3-(2-imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methyl-~{N}-[3-(4-methylpiperazin-1-yl)-5-(trifluoromethyl)phenyl]benzamide, NT-3 growth factor receptor | | Authors: | Zhang, Z.M, Wang, Y. | | Deposit date: | 2019-09-23 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis and biological evaluation of 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methylbenzamides as potent and selective pan-tropomyosin receptor kinase (TRK) inhibitors.
Eur.J.Med.Chem., 179, 2019
|
|
4DQE
 
 | | Crystal Structure of (G16C/L38C) HIV-1 Protease in Complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, Aspartyl protease | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-02-15 | | Release date: | 2012-03-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease.
J.Am.Chem.Soc., 134, 2012
|
|
3TT0
 
 | | Co-structure of Fibroblast Growth Factor Receptor 1 kinase domain with 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (BGJ398) | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-(6-{[4-(4-ethylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)-1-methylurea, Basic fibroblast growth factor receptor 1, GLYCEROL, ... | | Authors: | Bussiere, D.E, Murray, J.M, Shu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase.
J.Med.Chem., 54, 2011
|
|
5UG8
 
 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with a covalent inhibitor N-[(3R,4R)-4-fluoro-1-{6-[(1-methyl-1H-pyrazol-4-yl)amino]-9-(propan-2-yl)-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | Descriptor: | Epidermal growth factor receptor, GLYCEROL, N-[(3R,4R)-4-fluoro-1-{6-[(1-methyl-1H-pyrazol-4-yl)amino]-9-(propan-2-yl)-9H-purin-2-yl}pyrrolidin-3-yl]propanamide, ... | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2017-01-07 | | Release date: | 2017-03-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
4AIE
 
 | | Structure of glucan-1,6-alpha-glucosidase from Lactobacillus acidophilus NCFM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLUCAN 1,6-ALPHA-GLUCOSIDASE, ... | | Authors: | Fredslund, F, Navarro Poulsen, J.C, Lo Leggio, L. | | Deposit date: | 2012-02-09 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Enzymology and Structure of the Gh13_31 Glucan 1,6-Alpha-Glucosidase that Confers Isomaltooligosaccharide Utilization in the Probiotic Lactobacillus Acidophilus Ncfm.
J.Bacteriol., 194, 2012
|
|
6IVV
 
 | | Structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii with multiple surface binding regions at 1.26A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Viswanathan, V, Sharma, P, Chaudhary, A, Sharma, S, Singh, T.P. | | Deposit date: | 2018-12-04 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structure of peptide t-RNA hydrolase from Acinetobacter baumannii with multiple surface binding sites at 1.26 Angstrom resolution.
To Be Published
|
|
5D3G
 
 | | Structure of HIV-1 Reverse Transcriptase Bound to a Novel 38-mer Hairpin Template-Primer DNA Aptamer | | Descriptor: | DNA aptamer (38-MER), GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | Authors: | Miller, M.T, Tuske, S, Das, K, Arnold, E. | | Deposit date: | 2015-08-06 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of HIV-1 reverse transcriptase bound to a novel 38-mer hairpin template-primer DNA aptamer.
Protein Sci., 25, 2016
|
|
5GNK
 
 | | Crystal structure of EGFR 696-988 T790M in complex with LXX-6-34 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3R)-3-[4-azanyl-3-[3-chloranyl-4-[(1-methylimidazol-2-yl)methoxy]phenyl]pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]prop-2-en-1-one, CHLORIDE ION, ... | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2016-07-21 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Discovery of (R)-1-(3-(4-Amino-3-(3-chloro-4-(pyridin-2-ylmethoxy)phenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl)piperidin-1-yl)prop-2-en-1-one (CHMFL-EGFR-202) as a Novel Irreversible EGFR Mutant Kinase Inhibitor with a Distinct Binding Mode.
J. Med. Chem., 60, 2017
|
|
3LZU
 
 | | Crystal Structure of a Nelfinavir Resistant HIV-1 CRF01_AE Protease variant (N88S) in Complex with the Protease Inhibitor Darunavir. | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 protease | | Authors: | Schiffer, C.A, Bandaranayake, R.M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
4DQC
 
 | | Crystal Structure of (G16C/L38C) HIV-1 Protease in Complex with DRV | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Aspartyl protease, GLYCEROL, ... | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-02-15 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease.
J.Am.Chem.Soc., 134, 2012
|
|
6A34
 
 | |
7E6Q
 
 | | Crystal structure of influenza A virus neuraminidase N5 complexed with 4'-phenyl-1,2,3-triazolylated oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-acetamido-3-pentan-3-yloxy-5-(4-phenyl-1,2,3-triazol-1-yl)cyclohexene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, P.F, Babayemi, O.O, Li, C.N, Fu, L.F, Zhang, S.S, Qi, J.X, Lv, X, Li, X.B. | | Deposit date: | 2021-02-23 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design of 5'-substituted 1,2,3-triazolylated oseltamivir derivatives as potent influenza neuraminidase inhibitors.
Rsc Adv, 11, 2021
|
|
6AN2
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING D4TTP AT PH 7.5 | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA PRIMER (5'- D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*GP)-3'), DNA TEMPLATE (5'- D(*AP*TP*GP*AP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Das, K, Arnold, E. | | Deposit date: | 2017-08-11 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of HIV-1 reverse transcriptase/d4TTP complex: Novel DNA cross-linking site and pH-dependent conformational changes.
Protein Sci., 28, 2019
|
|
7VIH
 
 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with CBP-307 | | Descriptor: | 1-[[2-fluoranyl-4-[5-[4-(2-methylpropyl)phenyl]-1,2,4-oxadiazol-3-yl]phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3MWS
 
 | | Crystal Structure of Group N HIV-1 Protease | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, HIV-1 Protease | | Authors: | Sayer, J.M, Agniswamy, J, Weber, I.T, Louis, J.M. | | Deposit date: | 2010-05-06 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Autocatalytic maturation, physical/chemical properties, and crystal structure of group N HIV-1 protease: relevance to drug resistance.
Protein Sci., 19, 2010
|
|
3N3I
 
 | | Crystal Structure of G48V/C95F tethered HIV-1 Protease/Saquinavir complex | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Protease | | Authors: | Prashar, V, Bihani, S.C, Das, A, Rao, D.R, Hosur, M.V. | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Insights into the mechanism of drug resistance: X-ray structure analysis of G48V/C95F tethered HIV-1 protease dimer/saquinavir complex
Biochem.Biophys.Res.Commun., 396, 2010
|
|
3LZS
 
 | | Crystal Structure of HIV-1 CRF01_AE Protease in Complex with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 protease | | Authors: | Schiffer, C.A, Bandaranayake, R.M. | | Deposit date: | 2010-03-01 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
6ASW
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING D4TTP AT PH 9.0 | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA (27-MER), DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*(DDG))-3'), ... | | Authors: | Martinez, S.E, Das, K, Arnold, E. | | Deposit date: | 2017-08-25 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Structure of HIV-1 reverse transcriptase/d4TTP complex: Novel DNA cross-linking site and pH-dependent conformational changes.
Protein Sci., 28, 2019
|
|
6BFA
 
 | | Calcium-Dependent Protein Kinase 1 from Toxoplasma gondii (TgCDPK1) in complex with inhibitor UW1553 | | Descriptor: | 1-{4-amino-3-[6-(cyclopropyloxy)naphthalen-2-yl]-1H-pyrazolo[3,4-d]pyrimidin-1-yl}-2-methylpropan-2-ol, Calmodulin-domain protein kinase 1 | | Authors: | Merritt, E.A. | | Deposit date: | 2017-10-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Development of an Orally Available and Central Nervous System (CNS) Penetrant Toxoplasma gondii Calcium-Dependent Protein Kinase 1 (TgCDPK1) Inhibitor with Minimal Human Ether-a-go-go-Related Gene (hERG) Activity for the Treatment of Toxoplasmosis.
J. Med. Chem., 59, 2016
|
|
4NT2
 
 | | Crystal structure of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with lyso-sphingomyelin (d18:1) at 2.4 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(R)-{[(2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl]oxy}(hydroxy)phosphoryl]oxy}-N,N,N-trimethylethanaminium, SULFATE ION, ... | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
