2LWT
 
 | |
1HH9
 
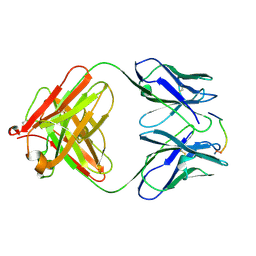 | | ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH A PEPTIDE | | Descriptor: | IGG2A KAPPA ANTIBODY CB41 (HEAVY CHAIN), IGG2A KAPPA ANTIBODY CB41 (LIGHT CHAIN), PEP-2 | | Authors: | Hahn, M, Wessner, H, Schneider-Mergener, J, Hohne, W. | | Deposit date: | 2000-12-21 | | Release date: | 2001-01-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Evolutionary Transition Pathways for Changing Peptide Ligand Specificity and Structure
Embo J., 19, 2000
|
|
2MLE
 
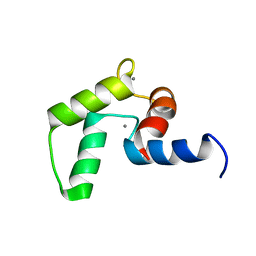 | | NMR structure of the C-domain of troponin C bound to the anchoring region of troponin I | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles | | Authors: | Robertson, I.M, Baryshnikova, O.K, Mercier, P, Sykes, B.D. | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dilated cardiomyopathy G159D mutation in cardiac troponin C weakens the anchoring interaction with troponin I.
Biochemistry, 47, 2008
|
|
1TMB
 
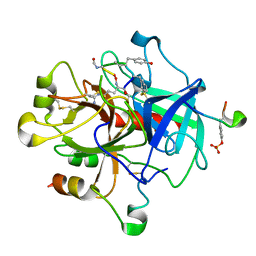 | | MOLECULAR BASIS FOR THE INHIBITION OF HUMAN ALPHA-THROMBIN BY THE MACROCYCLIC PEPTIDE CYCLOTHEONAMIDE A | | Descriptor: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), HIRUGEN, ... | | Authors: | Qiu, X, Padmanabhan, K.P, Maryanoff, B.E, Tulinsky, A. | | Deposit date: | 1993-05-27 | | Release date: | 1994-01-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the inhibition of human alpha-thrombin by the macrocyclic peptide cyclotheonamide A.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
2LWU
 
 | |
2LWS
 
 | |
2LWQ
 
 | |
1DQ9
 
 | | COMPLEX OF CATALYTIC PORTION OF HUMAN HMG-COA REDUCTASE WITH HMG-COA | | Descriptor: | 3-HYDROXY-3-METHYLGLUTARYL-COENZYME A, PROTEIN (HMG-COA REDUCTASE) | | Authors: | Istvan, E.S, Palnitkar, M, Buchanan, S.K, Deisenhofer, J. | | Deposit date: | 1999-12-30 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis.
EMBO J., 19, 2000
|
|
1HH6
 
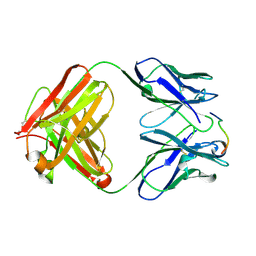 | | ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH A PEPTIDE | | Descriptor: | IGG2A KAPPA ANTIBODY CB41 (HEAVY CHAIN), IGG2A KAPPA ANTIBODY CB41 (LIGHT CHAIN), PEP-4 | | Authors: | Hahn, M, Wessner, H, Schneider-Mergener, J, Hohne, W. | | Deposit date: | 2000-12-21 | | Release date: | 2001-01-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolutionary Transition Pathways for Changing Peptide Ligand Specificity and Structure
Embo J., 19, 2000
|
|
1U5M
 
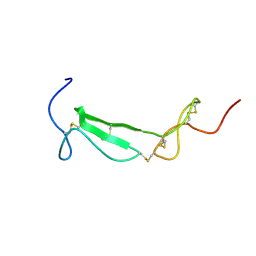 | | Structure of a Chordin-like Cysteine-rich Repeat (VWC module) from Collagen IIA | | Descriptor: | alpha 1 type II collagen isoform 1 | | Authors: | O'Leary, J.M, Hamilton, J.m, Deane, C.M, Valeyev, N.V, Sandell, L.J, Downing, A.K. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-05 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of a prototypical chordin-like cysteine-rich repeat (von Willebrand Factor type C module) from collagen IIA
J.Biol.Chem., 279, 2004
|
|
1Z32
 
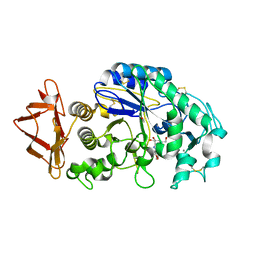 | | Structure-function relationships in human salivary alpha-amylase: Role of aromatic residues | | Descriptor: | 4-amino-4,6-dideoxy-alpha-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, CALCIUM ION, ... | | Authors: | Ramasubbu, N, Ragunath, C, Sundar, K, Mishra, P.J, Gyemant, G, Kandra, L. | | Deposit date: | 2005-03-10 | | Release date: | 2005-05-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-function relationships in human salivary alpha-amylase: Role of aromatic residues
To be Published
|
|
1UZI
 
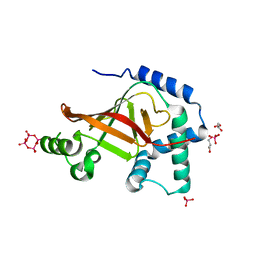 | | C3 EXOENZYME FROM CLOSTRIDIUM BOTULINUM, TETRAGONAL FORM | | Descriptor: | CYCLO-TETRAMETAVANADATE, GLYCEROL, MONO-ADP-RIBOSYLTRANSFERASE C3, ... | | Authors: | Evans, H.R, Holloway, D.E, Sutton, J.M, Ayriss, J, Shone, C.C, Acharya, K.R. | | Deposit date: | 2004-03-12 | | Release date: | 2004-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | C3 Exoenzyme from Clostridium Botulinum: Structure of a Tetragonal Crystal Form and a Reassessment of Nad-Induced Flexure
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1C28
 
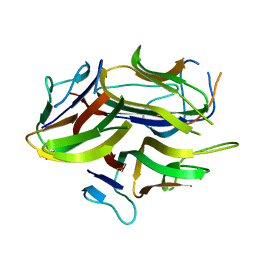 | |
1DSV
 
 | | STRUCTURE OF THE MMTV NUCLEOCAPSID PROTEIN (C-TERMINAL ZINC FINGER) | | Descriptor: | NUCLEIC ACID BINDING PROTEIN P14, ZINC ION | | Authors: | Klein, D.J, Johnson, P.E, Zollars, E.S, De Guzman, R.N, Summers, M.F. | | Deposit date: | 2000-01-08 | | Release date: | 2000-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle.
Biochemistry, 39, 2000
|
|
1DSQ
 
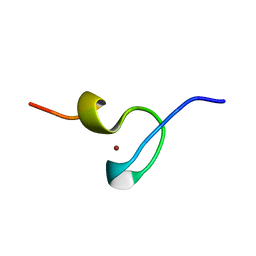 | | STRUCTURE OF THE MMTV NUCLEOCAPSID PROTEIN (ZINC FINGER 1) | | Descriptor: | NUCLEIC ACID BINDING PROTEIN P14, ZINC ION | | Authors: | Klein, D.J, Johnson, P.E, Zollars, E.S, De Guzman, R.N, Summers, M.F. | | Deposit date: | 2000-01-08 | | Release date: | 2000-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the nucleocapsid protein from the mouse mammary tumor virus reveals unusual folding of the C-terminal zinc knuckle.
Biochemistry, 39, 2000
|
|
1G27
 
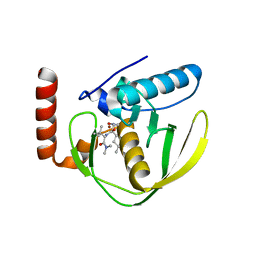 | | CRYSTAL STRUCTURE OF E.COLI POLYPEPTIDE DEFORMYLASE COMPLEXED WITH THE INHIBITOR BB-3497 | | Descriptor: | 2-[(FORMYL-HYDROXY-AMINO)-METHYL]-HEXANOIC ACID (1-DIMETHYLCARBAMOYL-2,2-DIMETHYL-PROPYL)-AMIDE, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | Authors: | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | Deposit date: | 2000-10-17 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
1G2A
 
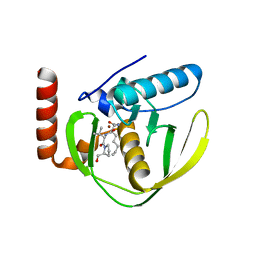 | | THE CRYSTAL STRUCTURE OF E.COLI PEPTIDE DEFORMYLASE COMPLEXED WITH ACTINONIN | | Descriptor: | ACTINONIN, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | Authors: | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | Deposit date: | 2000-10-18 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
5K1H
 
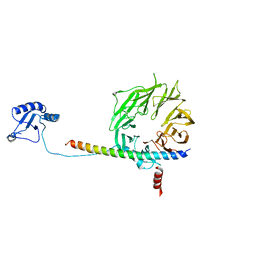 | | eIF3b relocated to the intersubunit face to interact with eIF1 and below the eIF2 ternary-complex. from the structure of a partial yeast 48S preinitiation complex in closed conformation. | | Descriptor: | Eukaryotic translation initiation factor 3 subunit B, eIF3a C-terminal tail | | Authors: | Simonetti, A, Brito Querido, J, Myasnikov, A.G, Mancera-Martinez, E, Renaud, A, Kuhn, L, Hashem, Y. | | Deposit date: | 2016-05-18 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | eIF3 Peripheral Subunits Rearrangement after mRNA Binding and Start-Codon Recognition.
Mol.Cell, 63, 2016
|
|
6I3M
 
 | | eIF2B:eIF2 complex, phosphorylated on eIF2 alpha serine 52. | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Adomavicius, T, Roseman, A.M, Pavitt, G.D. | | Deposit date: | 2018-11-06 | | Release date: | 2019-05-22 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | The structural basis of translational control by eIF2 phosphorylation.
Nat Commun, 10, 2019
|
|
6I7T
 
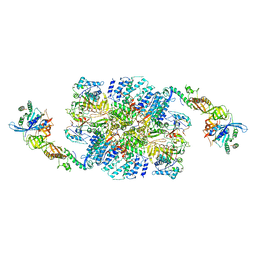 | | eIF2B:eIF2 complex | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Adomavicius, T, Guaita, M, Roseman, A.M, Pavitt, G.D. | | Deposit date: | 2018-11-17 | | Release date: | 2019-05-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.61 Å) | | Cite: | The structural basis of translational control by eIF2 phosphorylation.
Nat Commun, 10, 2019
|
|
5NVN
 
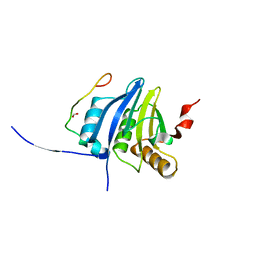 | | Crystal structure of the human 4EHP-4E-BP1 complex | | Descriptor: | Eukaryotic translation initiation factor 4E type 2, Eukaryotic translation initiation factor 4E-binding protein 1, FORMIC ACID | | Authors: | Peter, D, Sandmeir, F, Valkov, E. | | Deposit date: | 2017-05-04 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression.
Genes Dev., 31, 2017
|
|
5NVK
 
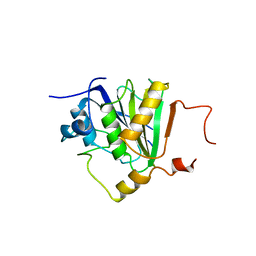 | | Crystal structure of the human 4EHP-GIGYF1 complex | | Descriptor: | Eukaryotic translation initiation factor 4E type 2, GRB10-interacting GYF protein 1 | | Authors: | Peter, D, Valkov, E. | | Deposit date: | 2017-05-04 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression.
Genes Dev., 31, 2017
|
|
5NVL
 
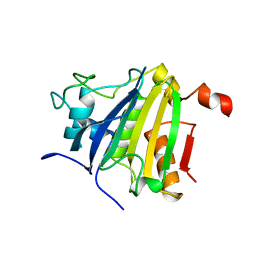 | | Crystal structure of the human 4EHP-GIGYF2 complex | | Descriptor: | Eukaryotic translation initiation factor 4E type 2, GRB10-interacting GYF protein 2 | | Authors: | Peter, D, Valkov, E. | | Deposit date: | 2017-05-04 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression.
Genes Dev., 31, 2017
|
|
1QCN
 
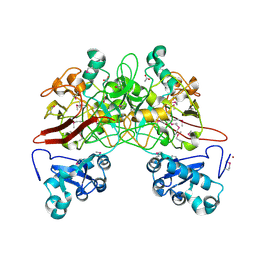 | | CRYSTAL STRUCTURE OF FUMARYLACETOACETATE HYDROLASE | | Descriptor: | ACETATE ION, CALCIUM ION, FUMARYLACETOACETATE HYDROLASE, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-05-14 | | Release date: | 2000-06-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
3BKQ
 
 | |
