8TJF
 
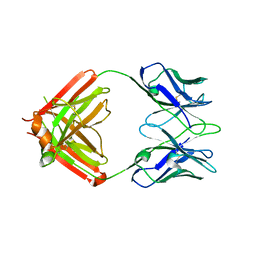 | | monovalent bispecific IgG antibodies through novel electrostatic steering mutations at the CH1-CL interface | | Descriptor: | Fab Lambda light chain, IgG1 Fab heavy chain | | Authors: | Oganesyan, V.Y, van Dyk, N, Mazor, Y, Chiang, C. | | Deposit date: | 2023-07-21 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Robust production of monovalent bispecific IgG antibodies through novel electrostatic steering mutations at the C H 1-C lambda interface.
Mabs, 15, 2023
|
|
7KQ3
 
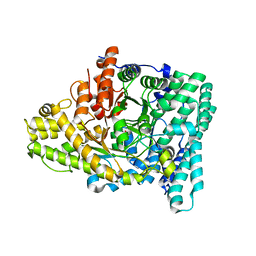 | |
3BJN
 
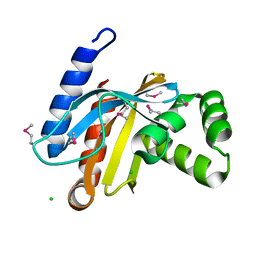 | | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae, targeted domain 79-240 | | Descriptor: | CHLORIDE ION, Transcriptional regulator, putative | | Authors: | Chang, C, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of C-terminal domain of putative transcriptional regulator from Vibrio cholerae.
To be Published
|
|
4FZJ
 
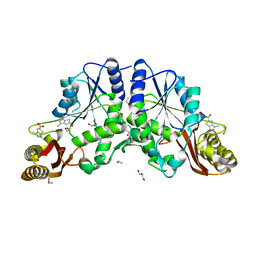 | | Pantothenate synthetase in complex with 1,3-DIMETHYL-1H-THIENO[2,3-C]PYRAZOLE-5-CARBOXYLIC ACID | | Descriptor: | 1,2-ETHANEDIOL, 1,3-dimethyl-1H-thieno[2,3-c]pyrazole-5-carboxylic acid, ETHANOL, ... | | Authors: | Silvestre, H.L, Blundell, T.L, Abell, C, Ciulli, A. | | Deposit date: | 2012-07-06 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Integrated biophysical approach to fragment screening and validation for fragment-based lead discovery.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1NNO
 
 | | CONFORMATIONAL CHANGES OCCURRING UPON NO BINDING IN NITRITE REDUCTASE FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | HEME C, HEME D, NITRIC OXIDE, ... | | Authors: | Nurizzo, D, Tegoni, M, Cambillau, C. | | Deposit date: | 1998-07-20 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Conformational changes occurring upon reduction and NO binding in nitrite reductase from Pseudomonas aeruginosa.
Biochemistry, 37, 1998
|
|
1OKS
 
 | | Crystal structure of the measles virus phosphoprotein XD domain | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Johansson, K, Bourhis, J.-M, Campanacci, V, Cambillau, C, Canard, B, Longhi, S. | | Deposit date: | 2003-07-29 | | Release date: | 2003-09-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Measles Virus Phosphoprotein Domain Responsible for the Induced Folding of the C-Terminal Domain of the Nucleoprotein
J.Biol.Chem., 278, 2003
|
|
4FZR
 
 | | Crystal Structure of SsfS6, Streptomyces sp. SF2575 glycosyltransferase | | Descriptor: | SsfS6 | | Authors: | Wang, F, Zhou, M, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-07-07 | | Release date: | 2012-07-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Crystal structure of SsfS6, the putative C-glycosyltransferase involved in SF2575 biosynthesis.
Proteins, 81, 2013
|
|
5EDH
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH n4c(C)n1c(nc(n1)CCc2nc(nn2C)N3CCCC3)c(c4)CC, micromolar IC50=0.0037753 | | Descriptor: | 8-ethyl-5-methyl-2-[2-(2-methyl-5-pyrrolidin-1-yl-1,2,4-triazol-3-yl)ethyl]-[1,2,4]triazolo[1,5-c]pyrimidine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Rudolph, M.G. | | Deposit date: | 2015-10-21 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A Real-World Perspective on Molecular Design.
J.Med.Chem., 59, 2016
|
|
3TCX
 
 | | Structure of Engineered Single Domain ICAM-1 D1 with High-Affinity aL Integrin I Domain of Native C-Terminal Helix Conformation | | Descriptor: | Integrin alpha-L, Intercellular adhesion molecule 1, MAGNESIUM ION | | Authors: | Kang, S, Kim, C.U, Gu, X, Owens, R.M, van Rijn, S.J, Boonyaleepun, V, Mao, Y, Springer, T.A, Jin, M.M. | | Deposit date: | 2011-08-09 | | Release date: | 2011-08-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of Engineered Single Domain ICAM-1 D1 with High-Affinity L Integrin I Domain of Native C-Terminal Helix Conformation
To be Published
|
|
7F94
 
 | | Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel with two conformationally different hemichannels | | Descriptor: | A C-terminal deletion mutant of gap junction alpha-1 protein (Cx43-M257) | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2021-07-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
3UU9
 
 | | Structure of the free TvNiRb form of Thioalkalivibrio nitratireducens cytochrome c nitrite reductase | | Descriptor: | CALCIUM ION, Eight-heme nitrite reductase, HEME C, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonova, T.V, Tikhonov, A.V, Dorovatovskii, P.V, Popov, V.O. | | Deposit date: | 2011-11-28 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3OA4
 
 | | CRYSTAL STRUCTURE OF hypothetical protein BH1468 from Bacillus halodurans C-125 | | Descriptor: | GLYCEROL, SULFATE ION, ZINC ION, ... | | Authors: | Malashkevich, V.N, Patskovsky, Y, Garrett, S, Foti, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-08-04 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | CRYSTAL STRUCTURE OF hypothetical protein BH1468 from Bacillus halodurans C-125
To be Published
|
|
3FAV
 
 | | Structure of the CFP10-ESAT6 complex from Mycobacterium tuberculosis | | Descriptor: | 6 kDa early secretory antigenic target, ESAT-6-like protein esxB, IMIDAZOLE, ... | | Authors: | Poulsen, C, Holton, S.J, Wilmanns, M, Song, Y.H. | | Deposit date: | 2008-11-18 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | WXG100 protein superfamily consists of three subfamilies and exhibits an alpha-helical C-terminal conserved residue pattern.
Plos One, 9, 2014
|
|
1KPE
 
 | | PKCI-TRANSITION STATE ANALOG | | Descriptor: | ADENOSINE-5'-DITUNGSTATE, PROTEIN KINASE C INTERACTING PROTEIN | | Authors: | Lima, C.D, Klein, M.G, Hendrickson, W.A. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based analysis of catalysis and substrate definition in the HIT protein family.
Science, 278, 1997
|
|
1YLG
 
 | | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor | | Descriptor: | apolipoprotein B mRNA | | Authors: | Maris, C, Masse, J, Allain, F.H, Chester, A, Navaratnam, N. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor.
Rna, 11, 2005
|
|
1WP0
 
 | | Human SCO1 | | Descriptor: | SCO1 protein homolog | | Authors: | Williams, J.C, Sue, C, Banting, G.S, Yang, H, Glerum, D.M, Hendrickson, W.A, Schon, E.A. | | Deposit date: | 2004-08-27 | | Release date: | 2005-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human SCO1: IMPLICATIONS FOR REDOX SIGNALING BY A MITOCHONDRIAL CYTOCHROME c OXIDASE "ASSEMBLY" PROTEIN
J.Biol.Chem., 280, 2005
|
|
2LFU
 
 | | The structure of a N. meningitides protein targeted for vaccine development | | Descriptor: | Gna2132 | | Authors: | Esposito, V, Musi, V, De Chiara, C, Kelly, G, Veggi, D, Pizza, M, Pastore, A. | | Deposit date: | 2011-07-15 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal Domain of Neisseria Heparin Binding Antigen (NHBA), One of the Main Antigens of a Novel Vaccine against Neisseria meningitidis.
J.Biol.Chem., 286, 2011
|
|
1U5M
 
 | | Structure of a Chordin-like Cysteine-rich Repeat (VWC module) from Collagen IIA | | Descriptor: | alpha 1 type II collagen isoform 1 | | Authors: | O'Leary, J.M, Hamilton, J.m, Deane, C.M, Valeyev, N.V, Sandell, L.J, Downing, A.K. | | Deposit date: | 2004-07-28 | | Release date: | 2004-10-05 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of a prototypical chordin-like cysteine-rich repeat (von Willebrand Factor type C module) from collagen IIA
J.Biol.Chem., 279, 2004
|
|
3Q9V
 
 | |
4IJ7
 
 | | Crystal structure of Odorant Binding Protein 48 from Anopheles gambiae (AgamOBP48) with PEG | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, Odorant binding protein-8, SODIUM ION | | Authors: | Zographos, S.E, Tsitsanou, K.E, Drakou, C.E. | | Deposit date: | 2012-12-21 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal and Solution Studies of the "Plus-C" Odorant-binding Protein 48 from Anopheles gambiae: CONTROL OF BINDING SPECIFICITY THROUGH THREE-DIMENSIONAL DOMAIN SWAPPING.
J.Biol.Chem., 288, 2013
|
|
3VDM
 
 | | Crystal Structure of VldE, the pseudo-glycosyltransferase which catalyzes non-glycosidic C-N coupling in Validamycin A biosynthesis | | Descriptor: | VldE | | Authors: | Cavalier, M.C, Yim, Y.-S, Asamizu, S, Neau, D, Mahmud, T, Lee, Y.-H. | | Deposit date: | 2012-01-05 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of VldE, the pseudo-glycosyltransferase which catalyzes non-glycosidic C-N coupling in Validamycin A biosynthesis
To be Published
|
|
4IG7
 
 | |
2LLH
 
 | | NMR structure of Npm1_c70 | | Descriptor: | Nucleophosmin | | Authors: | Banci, L, Bertini, I, Brunori, M, Di Matteo, A, Federici, L, Gallo, A, Lo Sterzo, C, Mori, M. | | Deposit date: | 2011-11-09 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of Nucleophosmin DNA-binding Domain and Analysis of Its Complex with a G-quadruplex Sequence from the c-MYC Promoter.
J.Biol.Chem., 287, 2012
|
|
1SBS
 
 | | CRYSTAL STRUCTURE OF AN ANTI-HCG FAB | | Descriptor: | MONOCLONAL ANTIBODY 3A2, SULFATE ION | | Authors: | Fotinou, C, Beauchamp, J, Emsley, P, Dehaan, A, Schielen, W.J.G, Bos, E, Isaacs, N.W. | | Deposit date: | 1998-04-08 | | Release date: | 1999-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of an Fab fragment against a C-terminal peptide of hCG at 2.0 A resolution.
J.Biol.Chem., 273, 1998
|
|
2K1S
 
 | | Solution NMR structure of the folded C-terminal fragment of YiaD from Escherichia coli. Northeast Structural Genomics Consortium target ER553. | | Descriptor: | Inner membrane lipoprotein yiaD | | Authors: | Ramelot, T.A, Zhao, L, Hamilton, K, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-14 | | Release date: | 2008-05-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the folded C-terminal fragment of YiaD from Escherichia coli. Northeast Structural Genomics Consortium target ER553.
To be Published
|
|
