2L4Z
 
 | | NMR structure of fusion of CtIP (641-685) to LMO4-LIM1 (18-82) | | Descriptor: | DNA endonuclease RBBP8,LIM domain transcription factor LMO4, ZINC ION | | Authors: | Liew, C, Stokes, P.H, Kwan, A.H, Matthews, J.M. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Interaction of the Breast Cancer Oncogene LMO4 with the Tumour Suppressor CtIP/RBBP8.
J.Mol.Biol., 425, 2013
|
|
2LEF
 
 | | LEF1 HMG DOMAIN (FROM MOUSE), COMPLEXED WITH DNA (15BP), NMR, 12 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*AP*CP*CP*CP*TP*TP*TP*GP*AP*AP*GP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*CP*TP*TP*CP*AP*AP*AP*GP*GP*GP*TP*G)-3'), PROTEIN (LYMPHOID ENHANCER-BINDING FACTOR) | | Authors: | Li, X, Love, J.J, Case, D.A, Wright, P.E. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for DNA bending by the architectural transcription factor LEF-1.
Nature, 376, 1995
|
|
2LJ9
 
 | |
2LJL
 
 | |
1E99
 
 | | Human thymidylate kinase complexed with AZTMP and ADP | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostermann, N, Lavie, A, Padiyar, S, Brundiers, R, Veit, T, Reinstein, J, Goody, R.S, Konrad, M, Schlichting, I. | | Deposit date: | 2000-10-10 | | Release date: | 2001-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Potentiating Azt Activation: Structures of Wildtype and Mutant Human Thymidylate Kinase Suggest Reasons for the Mutants' Improved Kinetics with the HIV Prodrug Metabolite Aztmp
J.Mol.Biol., 304, 2000
|
|
1E98
 
 | | Wild type human thymidylate kinase complexed with AZTMP and ADP | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostermann, N, Lavie, A, Padiyar, S, Brundiers, R, Veit, T, Reintein, J, Goody, R.S, Konrad, M, Schlichting, I. | | Deposit date: | 2000-10-10 | | Release date: | 2001-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Potentiating Azt Activation: Structures of Wildtype and Mutant Human Thymidylate Kinase Suggest Reasons for the Mutants' Improved Kinetics with the HIV Prodrug Metabolite Aztmp
J.Mol.Biol., 304, 2000
|
|
1E9B
 
 | | Human thymidylate kinase complexed with AZTMP and APPNP | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostermann, N, Lavie, A, Padiyar, S, Brundiers, R, Veit, T, Reintein, J, Goody, R.S, Konrad, M, Schlichting, I. | | Deposit date: | 2000-10-10 | | Release date: | 2001-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Potentiating Azt Activation: Structures of Wildtype and Mutant Human Thymidylate Kinase Suggest Reasons for the Mutants' Improved Kinetics with the HIV Prodrug Metabolite Aztmp
J.Mol.Biol., 304, 2000
|
|
3I6W
 
 | |
3I6U
 
 | |
1FP4
 
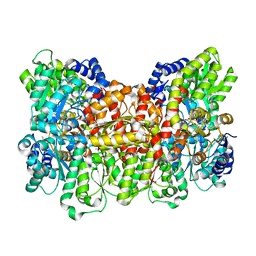 | | CRYSTAL STRUCTURE OF THE ALPHA-H195Q MUTANT OF NITROGENASE | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE-MO-S CLUSTER, ... | | Authors: | Sorlie, M, Christiansen, J, Lemon, B.J, Peters, J.W, Dean, D.R, Hales, B.J. | | Deposit date: | 2000-08-30 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic features and structure of the nitrogenase alpha-Gln195 MoFe protein
Biochemistry, 40, 2001
|
|
3J9T
 
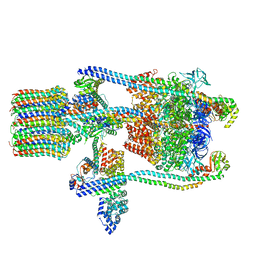 | | Yeast V-ATPase state 1 | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
3K2L
 
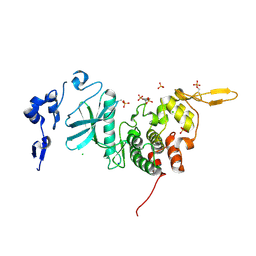 | | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) | | Descriptor: | CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 2, SODIUM ION, ... | | Authors: | Filippakopoulos, P, Myrianthopoulos, V, Soundararajan, M, Krojer, T, Hapka, E, Fedorov, O, Berridge, G, Wang, J, Shrestha, L, Pike, A.C.W, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Mikros, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structures of Down Syndrome Kinases, DYRKs, Reveal Mechanisms of Kinase Activation and Substrate Recognition.
Structure, 21, 2013
|
|
3J9V
 
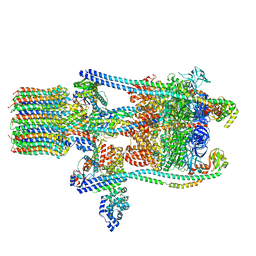 | | Yeast V-ATPase state 3 | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
3J9U
 
 | | Yeast V-ATPase state 2 | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
1GGW
 
 | |
3KVW
 
 | | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with an indirubin ligand | | Descriptor: | (2Z,3E)-7'-bromo-3-(hydroxyimino)-2'-oxo-1,1',2',3-tetrahydro-2,3'-biindole-5-carboxylic acid, CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Filippakopoulos, P, Myrianthopoulos, V, Kritsanida, M, Magiatis, P, Skaltsounis, A.L, Soundararajan, M, Krojer, T, Gileadi, O, Hapka, E, Fedorov, O, Berridge, G, Wang, J, Shrestha, L, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Mikros, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-11-30 | | Release date: | 2010-01-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) in complex with an indirubin ligand
To be Published
|
|
1H6O
 
 | | Dimerisation domain from human TRF1 | | Descriptor: | TELOMERIC REPEAT BINDING FACTOR 1 | | Authors: | Fairall, L, Chapman, L, Rhodes, D. | | Deposit date: | 2001-06-20 | | Release date: | 2001-09-05 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2.
Mol.Cell, 8, 2001
|
|
1H6P
 
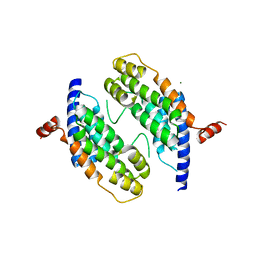 | | Dimeristion domain from human TRF2 | | Descriptor: | MAGNESIUM ION, TELOMERIC REPEAT BINDING FACTOR 2 | | Authors: | Chapman, L, Fairall, L, Rhodes, D. | | Deposit date: | 2001-06-20 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Trfh Dimerization Domain of the Human Telomere Proteins Trf1 and Trf2
Mol.Cell, 8, 2001
|
|
1HS1
 
 | |
1HS3
 
 | |
1HS8
 
 | |
1HS2
 
 | |
1HS4
 
 | |
3L82
 
 | | X-ray Crystal structure of TRF1 and Fbx4 complex | | Descriptor: | F-box only protein 4, Telomeric repeat-binding factor 1 | | Authors: | Zeng, Z.X, Wang, W, Yang, Y.T, Chen, Y, Yang, X.M, Diehl, J.A, Liu, X.D, Lei, M. | | Deposit date: | 2009-12-29 | | Release date: | 2010-03-09 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Selective Ubiquitination of TRF1 by SCF(Fbx4)
Dev.Cell, 18, 2010
|
|
6RML
 
 | | Crystal structure of TOPBP1 BRCT0,1,2 in complex with a 53BP1 phosphopeptide | | Descriptor: | 53BP1, DNA topoisomerase 2-binding protein 1 | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2019-05-07 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Phosphorylation-mediated interactions with TOPBP1 couple 53BP1 and 9-1-1 to control the G1 DNA damage checkpoint.
Elife, 8, 2019
|
|
