6U07
 
 | | Computational Stabilization of T Cell Receptor Constant Domains | | Descriptor: | MAGNESIUM ION, Stabilized T cell receptor constant domain (Calpha), Stabilized T cell receptor constant domain (Cbeta) | | Authors: | Froning, K, Maguire, J, Sereno, A, Huang, F, Chang, S, Weichert, K, Frommelt, A.J, Dong, J, Wu, X, Austin, H, Conner, E.M, Fitchett, J.R, Heng, A.R, Balasubramaniam, D, Hilgers, M.T, Kuhlman, B, Demarest, S.J. | | Deposit date: | 2019-08-13 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Computational stabilization of T cell receptors allows pairing with antibodies to form bispecifics.
Nat Commun, 11, 2020
|
|
6OOG
 
 | |
1J9A
 
 | | OLIGORIBONUCLEASE | | Descriptor: | OLIGORIBONUCLEASE, SULFATE ION | | Authors: | Bonander, N, Tordova, M, Ladner, J.E, Eisenstein, E, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-24 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of Haemophilus Influenzae HI1715, an Oligoribonuclease
To be Published
|
|
8CN0
 
 | | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-21 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
8CNB
 
 | | Crystal structure of CREBBP-Y1503C histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Crystal structure of CREBBP-Y1503C histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
4Q4F
 
 | | Crystal structure of LIMP-2 (space group C2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-beta-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Ren, J, Padilla-Parra, S, Fry, L.E, Stuart, D.I. | | Deposit date: | 2014-04-14 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lysosome sorting of beta-glucocerebrosidase by LIMP-2 is targeted by the mannose 6-phosphate receptor.
Nat Commun, 5, 2014
|
|
7AWZ
 
 | |
8CMV
 
 | |
6OFZ
 
 | | Crystal structure of human WDR5 | | Descriptor: | WD repeat-containing protein 5 | | Authors: | Lorton, B.M, Harijan, R.K, Burgos, E, Bonanno, J.B, Almo, S.C, Shechter, D. | | Deposit date: | 2019-04-01 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Binary Arginine Methylation Switch on Histone H3 Arginine 2 Regulates Its Interaction with WDR5.
Biochemistry, 59, 2020
|
|
8CNA
 
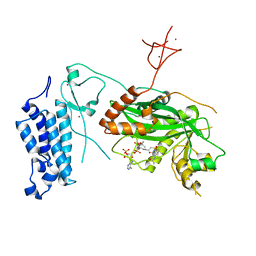 | | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | ZINC ION, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.463 Å) | | Cite: | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA
To Be Published
|
|
8CMZ
 
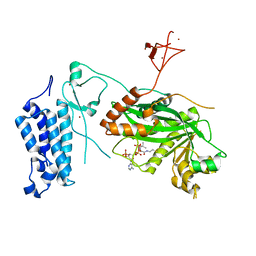 | | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-21 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
8CND
 
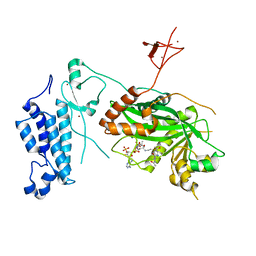 | | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | ZINC ION, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Crystal structure of CREBBP-Y1482N histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA
To Be Published
|
|
6EBZ
 
 | |
6EE5
 
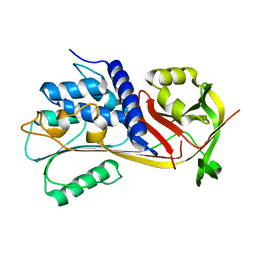 | |
3VBF
 
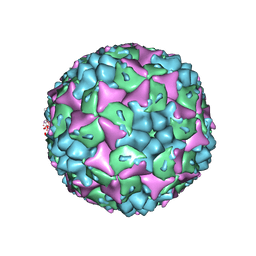 | | Crystal structure of formaldehyde treated human Enterovirus 71 (space group I23) | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Genome Polyprotein, ... | | Authors: | Wang, X, Peng, W, Ren, J, Hu, Z, Xu, J, Lou, Z, Li, X, Yin, W, Shen, X, Porta, C, Walter, T.S, Evans, G, Axford, D, Owen, R, Rowlands, D.J, Wang, J, Stuart, D.I, Fry, E.E, Rao, Z. | | Deposit date: | 2012-01-02 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71.
Nat.Struct.Mol.Biol., 19, 2012
|
|
8CQH
 
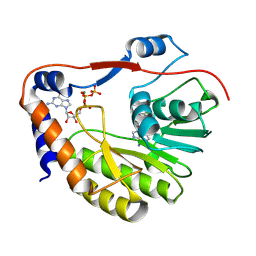 | |
4CVJ
 
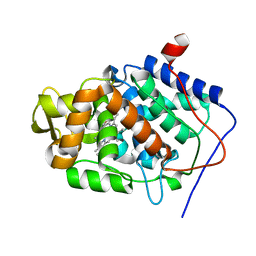 | | Neutron Structure of Compound I intermediate of Cytochrome c Peroxidase - Deuterium exchanged 100 K | | Descriptor: | CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Casadei, C.M, Gumiero, A, Blakeley, M.P, Ostermann, A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (2.182 Å), X-RAY DIFFRACTION | | Cite: | Neutron Cryo-Crystallography Captures the Protonation State of Ferryl Heme in a Peroxidase
Science, 345, 2014
|
|
5AN1
 
 | |
8C1Y
 
 | | Small molecule stabilizer for 14-3-3/ChREBP (Cmd 30) | | Descriptor: | 14-3-3 protein sigma, Carbohydrate-responsive element-binding protein, [2-[2-[[2,2-bis(fluoranyl)-2-phenyl-ethyl]amino]-2-oxidanylidene-ethoxy]phenyl]phosphonic acid | | Authors: | Pennings, M.A.M, Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-21 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular glues of the regulatory ChREBP/14-3-3 complex protect beta cells from glucolipotoxicity.
Biorxiv, 2024
|
|
8CHA
 
 | | Fc gamma RIIa 27W/131H variant ectodomain | | Descriptor: | 1,2-DIMETHOXYETHANE, 2-[2-(2-ethoxyethoxy)ethoxy]ethanol, 3,6,9,12,15,18-HEXAOXAICOSANE, ... | | Authors: | Foy, E.G, Thomsen, M, Goldman, A, Robinson, J.I. | | Deposit date: | 2023-02-07 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fc gamma RIIa 27W/131H variant ectodomain
To Be Published
|
|
1IVA
 
 | |
7SO1
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (E)-4-((4-((4-(2-cyanovinyl)-2,6-dimethylphenyl)amino)-6-(3-morpholinopropoxy)-1,3,5-triazin-2-yl)amino)benzonitrile (JLJ564) | | Descriptor: | 4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylanilino}-6-[3-(morpholin-4-yl)propoxy]-1,3,5-triazin-2-yl)amino]benzonitrile, Reverse transcriptase p66, p51 RT | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2021-10-28 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.727 Å) | | Cite: | Structural Studies and Structure Activity Relationships for Novel Computationally Designed Non-nucleoside Inhibitors and Their Interactions With HIV-1 Reverse Transcriptase.
Front Mol Biosci, 9, 2022
|
|
6UGT
 
 | | Crystal structure of the Fab fragment of PF06438179/GP1111 an infliximab biosimilar in a I-centered orthorhombic crystal form, Lot A | | Descriptor: | 1,2-ETHANEDIOL, PF06438179 Fab Heavy Chain, PF06438179 Fab Light Chain | | Authors: | Lerch, T.F, Sharpe, P, Mayclin, S.J, Edwards, T.E, Polleck, S, Rouse, J.C, Conlan, H.D. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
8BZG
 
 | | FC-31 stabilizer of 14-3-3 and ERalpha | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, Fusicoccin 31, ... | | Authors: | Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Stabilization of the Estrogen receptor alpha - 14-3-3 interaction as a potential intervention strategy for endocrine resistance in breast cancer
To Be Published
|
|
4D2P
 
 | | Structure of MELK in complex with inhibitors | | Descriptor: | 7-({4-[(3-hydroxy-5-methoxyphenyl)amino]benzoyl}amino)-1,2,3,4-tetrahydroisoquinolinium, MATERNAL EMBRYONIC LEUCINE ZIPPER KINASE | | Authors: | Johnson, C.N, Berdini, V, Beke, L, Bonnet, P, Brehmer, D, Coyle, J.E, Day, P.J, Frederickson, M, Freyne, E.J.E, Gilissen, R.A.H.J, Hamlett, C.C.F, Howard, S, Meerpoel, L, McMenamin, R, Patel, S, Rees, D.C, Sharff, A, Sommen, F, Wu, T, Linders, J.T.M. | | Deposit date: | 2014-05-12 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Fragment-Based Discovery of Type I Inhibitors of Maternal Embryonic Leucine Zipper Kinase
Acs Med.Chem.Lett., 6, 2015
|
|
