1MV5
 
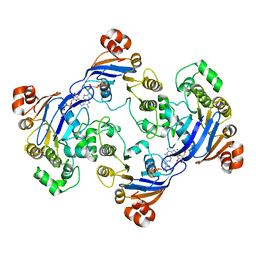 | | Crystal structure of LmrA ATP-binding domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yuan, Y, Chen, H, Patel, D. | | Deposit date: | 2002-09-24 | | Release date: | 2003-12-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of LmrA ATP-binding domain reveals the two-site alternating mechanism at molecular level
To be Published
|
|
2WAX
 
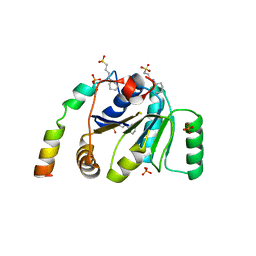 | |
7TR8
 
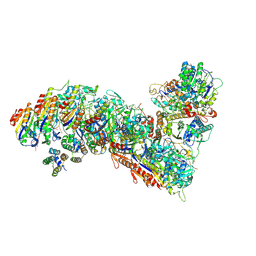 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
7TRA
 
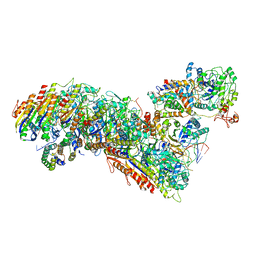 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
7TR9
 
 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
7TR6
 
 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | Cas11a, Cas5a, Cas7a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
8BXA
 
 | | Crystal structure of ribosome binding factor A (RbfA) from S. aureus | | Descriptor: | Ribosome-binding factor A | | Authors: | Fatkhullin, B, Bikmullin, A, Gabdulkhakov, A, Khusainov, I, Validov, S, Usachev, K, Yusupov, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Yet Another Similarity between Mitochondrial and Bacterial Ribosomal Small Subunit Biogenesis Obtained by Structural Characterization of RbfA from S. aureus.
Int J Mol Sci, 24, 2023
|
|
3IZZ
 
 | | Models for ribosome components that are nearest neighbors to the bovine mitochondrial initiation factor2 bound to the E. Coli ribosome | | Descriptor: | Helix 18 (Small Subunit), Helix 44 (Small Subunit), Helix 5, ... | | Authors: | Yassin, A.S, Haque, E, Datta, P.P, Elmore, K, Banavali, N.K, Spremulli, L.L, Agrawal, R.K. | | Deposit date: | 2011-01-20 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Insertion domain within mammalian mitochondrial translation initiation factor 2 serves the role of eubacterial initiation factor 1.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7ZO1
 
 | | SpCas9 bound to CD34 off-target9 DNA substrate | | Descriptor: | CD34 off-target9 DNA non-target strand, CD34 off-target9 DNA target strand, CD34 sgRNA, ... | | Authors: | Pacesa, M, Jinek, M. | | Deposit date: | 2022-04-23 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
6NBU
 
 | |
5V8I
 
 | | Thermus thermophilus 70S ribosome lacking ribosomal protein uS17 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Gregory, S.T, Jogl, G. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural robustness of the ribosome inferred from X-ray crystal structures of the 30S ribosomal subunit and the 70S ribosome lacking ribosomal protein uS17
To be published
|
|
3I5K
 
 | | Crystal structure of the NS5B polymerase from Hepatitis C Virus (HCV) strain JFH1 | | Descriptor: | PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Simister, P.C, Schmitt, M, Lohmann, V, Bressanelli, S. | | Deposit date: | 2009-07-05 | | Release date: | 2009-09-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of hepatitis C virus strain JFH1 polymerase
J.Virol., 83, 2009
|
|
2WAY
 
 | |
5E7M
 
 | |
4EQK
 
 | |
4ENF
 
 | | Crystal structure of the cap-binding domain of polymerase basic protein 2 from influenza virus A/Puerto Rico/8/34(h1n1) | | Descriptor: | 1,4-BUTANEDIOL, NITRATE ION, Polymerase basic protein 2 | | Authors: | Meng, G, Liu, Y, Zheng, X. | | Deposit date: | 2012-04-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural and functional characterization of K339T substitution identified in the PB2 subunit cap-binding pocket of influenza A virus
J.Biol.Chem., 288, 2013
|
|
4C0O
 
 | |
7F75
 
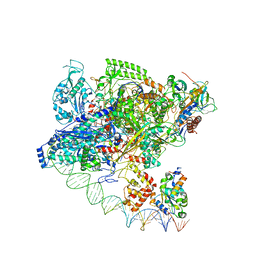 | | Cryo-EM structure of Spx-dependent transcription activation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Feng, Y, Shi, J. | | Deposit date: | 2021-06-28 | | Release date: | 2021-10-13 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of transcription activation by the global regulator Spx.
Nucleic Acids Res., 49, 2021
|
|
8G5P
 
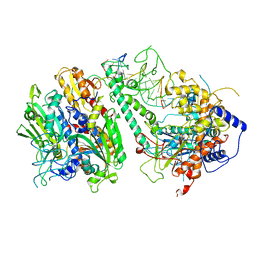 | | Cryo-EM structure of the Guide loop Engagement Complex (V) of Human Mitochondrial DNA Polymerase Gamma | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Nayak, A.R, Buchel, G, Herbine, K.H, Sarfallah, A, Sokolova, V.O, Zamudio-Ochoa, A, Temiakov, D. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural basis for DNA proofreading.
Nat Commun, 14, 2023
|
|
7FBR
 
 | |
8APU
 
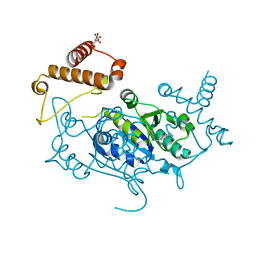 | | Crystal Structure of H. influenzae TrmD in complex with Compound 14 | | Descriptor: | 6-[[4-(aminomethyl)phenyl]methylamino]pyridine-3-carboxamide, CITRIC ACID, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Hall, G, Cowan, R, Carr, M.D. | | Deposit date: | 2022-08-10 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evaluating the druggability of TrmD, a potential antibacterial target, through design and microbiological profiling of a series of potent TrmD inhibitors.
Bioorg.Med.Chem.Lett., 90, 2023
|
|
8APT
 
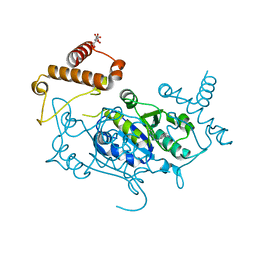 | | Crystal Structure of H. influenzae TrmD in complex with Compound 13 | | Descriptor: | 6-[[3-(aminomethyl)phenyl]methylamino]pyridine-3-carboxamide, CITRIC ACID, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Hall, G, Cowan, R, Carr, M.D. | | Deposit date: | 2022-08-10 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluating the druggability of TrmD, a potential antibacterial target, through design and microbiological profiling of a series of potent TrmD inhibitors.
Bioorg.Med.Chem.Lett., 90, 2023
|
|
8APV
 
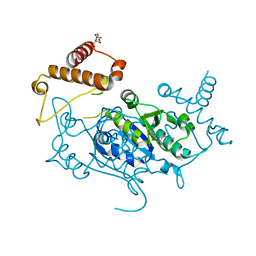 | | Crystal Structure of H. influenzae TrmD in complex with Compound 27 | | Descriptor: | 1-[[4-(aminomethyl)phenyl]methyl]pyrrolo[2,3-b]pyridine-5-carboxamide, CITRIC ACID, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Hall, G, Cowan, R, Carr, M.D. | | Deposit date: | 2022-08-10 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evaluating the druggability of TrmD, a potential antibacterial target, through design and microbiological profiling of a series of potent TrmD inhibitors.
Bioorg.Med.Chem.Lett., 90, 2023
|
|
8APW
 
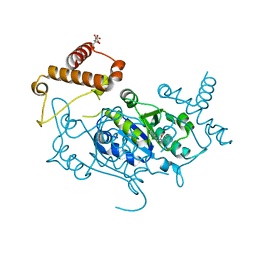 | | Crystal Structure of H. influenzae TrmD in complex with Compound 30 | | Descriptor: | 1-[2-oxidanylidene-2-(piperidin-4-ylamino)ethyl]pyrrolo[2,3-b]pyridine-5-carboxamide, CITRIC ACID, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Hall, G, Cowan, R, Carr, M.D. | | Deposit date: | 2022-08-10 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluating the druggability of TrmD, a potential antibacterial target, through design and microbiological profiling of a series of potent TrmD inhibitors.
Bioorg.Med.Chem.Lett., 90, 2023
|
|
5O60
 
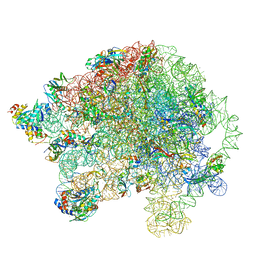 | | Structure of the 50S large ribosomal subunit from Mycobacterium smegmatis | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Hentschel, J, Burnside, C, Mignot, I, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2017-06-02 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The Complete Structure of the Mycobacterium smegmatis 70S Ribosome.
Cell Rep, 20, 2017
|
|
