3FEX
 
 | |
5W6Z
 
 | | Crystal structure of the H24W mutant of HsNUDT16 | | Descriptor: | SODIUM ION, U8 snoRNA-decapping enzyme | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
3VSD
 
 | | Crystal Structure of the K127A Mutant of O-Phosphoserine Sulfhydrylase Complexed with External Schiff Base of Pyridoxal 5'-Phosphate with O-Acetyl-L-Serine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, O-ACETYLSERINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nakamura, T, Kawai, Y, Kataoka, M, Ishikawa, K. | | Deposit date: | 2012-04-24 | | Release date: | 2012-05-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural analysis of the substrate recognition mechanism in O-phosphoserine sulfhydrylase from the hyperthermophilic archaeon Aeropyrum pernix K1
J.Mol.Biol., 422, 2012
|
|
8GXB
 
 | |
7O2I
 
 | | METTL3-METTL14 heterodimer bound to the SAM competitive small molecule inhibitor STM2457 | | Descriptor: | DIMETHYL SULFOXIDE, N6-adenosine-methyltransferase catalytic subunit, N6-adenosine-methyltransferase non-catalytic subunit, ... | | Authors: | Pilka, E.S, Blackaby, W, Hardick, D, Harper, C, Hewstone, D, Ridgill, M, Rotty, B, Rausch, O. | | Deposit date: | 2021-03-30 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia.
Nature, 593, 2021
|
|
6MVQ
 
 | | HCV NS5B 1b N316 bound to Compound 31 | | Descriptor: | (4-{1-[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl]-1H-1,2,4-triazol-5-yl}-2-fluorophenyl)boronic acid, HCV Polymerase | | Authors: | Williams, S.P, Kahler, K, Price, D.J, Peat, A.J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Design of N-Benzoxaborole Benzofuran GSK8175-Optimization of Human Pharmacokinetics Inspired by Metabolites of a Failed Clinical HCV Inhibitor.
J.Med.Chem., 62, 2019
|
|
6MVO
 
 | | HCV NS5B 1A Y316 bound to Compound 49 | | Descriptor: | 6-[(7-chloro-1-hydroxy-1,3-dihydro-2,1-benzoxaborol-5-yl)(methylsulfonyl)amino]-5-cyclopropyl-2-(4-fluorophenyl)-N-methyl-1-benzofuran-3-carboxamide, RNA-directed RNA polymerase, SULFATE ION | | Authors: | Williams, S.P, Kahler, K, Price, D.J, Peat, A.J. | | Deposit date: | 2018-10-26 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design of N-Benzoxaborole Benzofuran GSK8175-Optimization of Human Pharmacokinetics Inspired by Metabolites of a Failed Clinical HCV Inhibitor.
J.Med.Chem., 62, 2019
|
|
5N65
 
 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9h | | Descriptor: | 2-phenyl-~{N}4-(2-thiophen-2-ylethyl)quinazoline-4,7-diamine, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
5N64
 
 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9g | | Descriptor: | 2-phenyl-~{N}4-(thiophen-2-ylmethyl)quinazoline-4,7-diamine, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
5N66
 
 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9j | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Mitogen-activated protein kinase 14, ~{N}4-[[4-(cyclopropylmethyl)furan-2-yl]methyl]-2-phenyl-quinazoline-4,7-diamine | | Authors: | Buehrmann, M, Mueller, M.P, Wiedemann, B, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
5N67
 
 | |
5N68
 
 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9m | | Descriptor: | 2-(4-morpholin-4-ylphenyl)-~{N}4-(2-phenylethyl)quinazoline-4,7-diamine, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
5N63
 
 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9c | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}4-[(4-fluorophenyl)methyl]-2-phenyl-quinazoline-4,7-diamine | | Authors: | Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
8FOK
 
 | | Cryo-EM structure of S. cerevisiae DNA polymerase alpha-primase complex in the DNA elongation state | | Descriptor: | DNA polymerase, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Yuan, Z, Georgescu, R, Li, H, O'Donnell, M. | | Deposit date: | 2022-12-30 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Molecular choreography of primer synthesis by the eukaryotic Pol alpha-primase.
Nat Commun, 14, 2023
|
|
6K6U
 
 | |
2FTC
 
 | | Structural Model for the Large Subunit of the Mammalian Mitochondrial Ribosome | | Descriptor: | 39S ribosomal protein L11, mitochondrial, 39S ribosomal protein L12, ... | | Authors: | Mears, J.A, Sharma, M.R, Gutell, R.R, Richardson, P.E, Agrawal, R.K, Harvey, S.C. | | Deposit date: | 2006-01-24 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.1 Å) | | Cite: | A Structural Model for the Large Subunit of the Mammalian Mitochondrial Ribosome
J.Mol.Biol., 358, 2006
|
|
5VYQ
 
 | | Crystal structure of the N-formyltransferase Rv3404c from mycobacterium tuberculosis in complex with YDP-Qui4N and folinic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Dunsirn, M.M, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-05-26 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical Investigation of Rv3404c from Mycobacterium tuberculosis.
Biochemistry, 56, 2017
|
|
6V3D
 
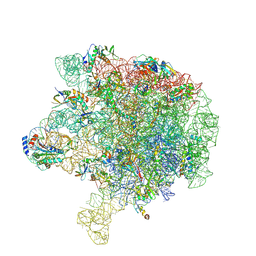 | |
7R98
 
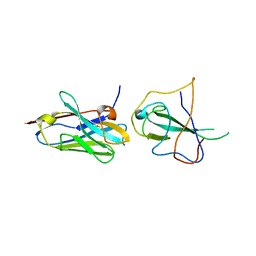 | |
8FRK
 
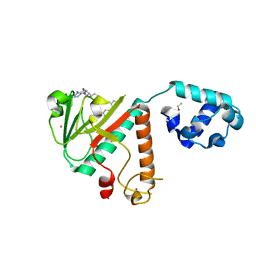 | |
8FRJ
 
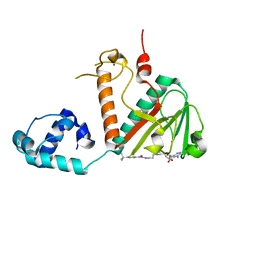 | | Structure of nsp14 N7-MethylTransferase domain fused with TELSAM bound to SGC0946 | | Descriptor: | 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Transcription factor ETV6,Guanine-N7 methyltransferase nsp14 chimera, ZINC ION | | Authors: | Kottur, J, Aggarwal, A.K. | | Deposit date: | 2023-01-07 | | Release date: | 2023-07-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structures of SARS-CoV-2 N7-methyltransferase with DOT1L and PRMT7 inhibitors provide a platform for new antivirals.
Plos Pathog., 19, 2023
|
|
5LYU
 
 | | The native crystal structure of 7SK 5'-hairpin | | Descriptor: | 7SK RNA, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Martinez-Zapien, D, Legrand, P, McEwen, A.C, Pasquali, S, Dock-Bregeon, A.-C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the 5 functional domain of the transcription riboregulator 7SK.
Nucleic Acids Res., 45, 2017
|
|
6S12
 
 | | Erythromycin Resistant Staphylococcus aureus 50S ribosome (delta R88 A89 uL22). | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Halfon, Y, Matozv, D, Eyal, Z, Bashan, A, Zimmerman, E, Kjeldgaard, J, Ingmer, H, Yonath, A. | | Deposit date: | 2019-06-18 | | Release date: | 2019-08-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Exit tunnel modulation as resistance mechanism of S. aureus erythromycin resistant mutant.
Sci Rep, 9, 2019
|
|
5LYV
 
 | | The crystal structure of 7SK 5'-hairpin - Osmium derivative | | Descriptor: | 7SK RNA, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Martinez-Zapien, D, Legrand, P, McEwen, A.G, Pasquali, S, Dock-Bregeon, A.-C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of the 5 functional domain of the transcription riboregulator 7SK.
Nucleic Acids Res., 45, 2017
|
|
7ML8
 
 | | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ001023038 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-2-phenyl-1,6-dihydropyrimidine-4-carboxamide, GLUTAMIC ACID, Hexa Vinylpyrrolidone K15, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Yun, M.K, Dubois, R, Rankovic, Z, White, S.W. | | Deposit date: | 2021-04-27 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of I38T mutant PA endonuclease (2009/H1N1/CALIFORNIA) in complex with St Jude compound AC067-19
To Be Published
|
|
