3M93
 
 | |
2VDN
 
 | | Re-refinement of Integrin AlphaIIbBeta3 Headpiece Bound to Antagonist Eptifibatide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, EPTIFIBATIDE, ... | | Authors: | Springer, T.A, Zhu, J, Xiao, T. | | Deposit date: | 2007-10-10 | | Release date: | 2008-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Distinctive Recognition of Fibrinogen Gammac Peptide by the Platelet Integrin Alphaiibbeta3.
J.Cell Biol., 182, 2008
|
|
2XBU
 
 | | Saccharomyces cerevisiae hypoxanthine-guanine phosphoribosyltransferase in complex with GMP (monoclinic crystal form) | | Descriptor: | ACETATE ION, GUANOSINE-5'-MONOPHOSPHATE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, ... | | Authors: | Moynie, L, Giraud, M.F, Breton, A, Boissier, F, Daignan-Fornier, B, Dautant, A. | | Deposit date: | 2010-04-15 | | Release date: | 2011-05-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional Significance of Four Successive Glycine Residues in the Pyrophosphate Binding Loop of Fungal 6-Oxopurine Phosphoribosyltransferases.
Protein Sci., 21, 2012
|
|
8OWX
 
 | | Crystal Structure of METTL6 bound to SAH | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA N(3)-methylcytidine methyltransferase METTL6 | | Authors: | Throll, P, Basu, S, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 2024
|
|
8OWY
 
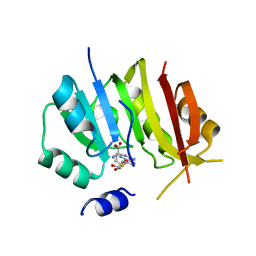 | |
1W2M
 
 | | Ca-substituted form of E. coli aminopeptidase P | | Descriptor: | CALCIUM ION, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Graham, S.C, Bond, C.S, Freeman, H.C, Guss, J.M. | | Deposit date: | 2004-07-07 | | Release date: | 2005-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Functional Implications of Metal Ion Selection in Aminopeptidase P, a Metalloprotease with a Dinuclear Metal Center.
Biochemistry, 44, 2005
|
|
2VMD
 
 | | Structure of the complex of discoidin II from Dictyostelium discoideum with beta-methyl-galactose | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, CALCIUM ION, ... | | Authors: | Aragao, K.S, Satre, M, Imberty, A, Varrot, A. | | Deposit date: | 2008-01-25 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure Determination of Discoidin II from Dictyostelium Discoideum and Carbohydrate Binding Properties of the Lectin Domain.
Proteins, 73, 2008
|
|
8OUH
 
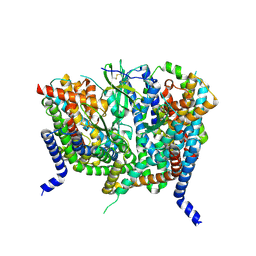 | | Complex of human ASCT2 with Syncytin-1 | | Descriptor: | ALANINE, Neutral amino acid transporter B(0), Syncytin-1 | | Authors: | Khare, S, Reyes, N. | | Deposit date: | 2023-04-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Receptor-recognition and antiviral mechanisms of retrovirus-derived human proteins.
Nat.Struct.Mol.Biol., 2024
|
|
2V8M
 
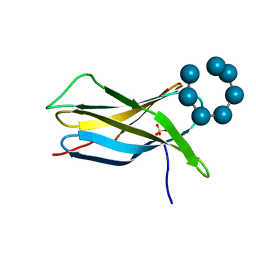 | |
3MEC
 
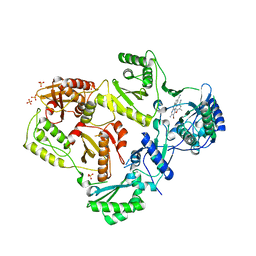 | | HIV-1 Reverse Transcriptase in Complex with TMC125 | | Descriptor: | 4-({6-AMINO-5-BROMO-2-[(4-CYANOPHENYL)AMINO]PYRIMIDIN-4-YL}OXY)-3,5-DIMETHYLBENZONITRILE, SULFATE ION, p51 Reverse transcriptase, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase with Etravirine (TMC125) and Rilpivirine (TMC278): Implications for Drug Design.
J.Med.Chem., 53, 2010
|
|
1W1U
 
 | |
8PIH
 
 | | Structure of Api m1 in complex with two nanobodies | | Descriptor: | CHLORIDE ION, Phospholipase A2, nanobdoy AM1-1, ... | | Authors: | Aagaard, J.B, Gandini, R, Spillner, E, Miehe, M, Andersen, G.R. | | Deposit date: | 2023-06-21 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Nanobody-based IgG formats as blocking antibodies of the major honeybee venom allergen Api m 1
To be published
|
|
2VG2
 
 | | Rv2361 with IPP | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CHLORIDE ION, DIPHOSPHATE, ... | | Authors: | Naismith, J.H, Wang, W, Dong, C. | | Deposit date: | 2007-11-07 | | Release date: | 2007-11-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structural basis of chain length control in Rv1086.
J. Mol. Biol., 381, 2008
|
|
6K5F
 
 | | Crystal structure of the CLC-ec1 deltaNC in presence of 200 mM NaBr | | Descriptor: | BROMIDE ION, Fab fragment, heavy chain, ... | | Authors: | Park, K, Lim, H.H. | | Deposit date: | 2019-05-28 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | Mutation of external glutamate residue reveals a new intermediate transport state and anion binding site in a CLC Cl-/H+antiporter.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3MPJ
 
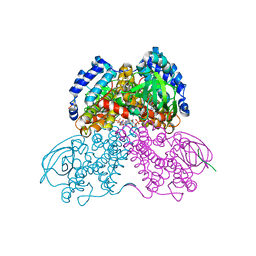 | | Structure of the glutaryl-coenzyme A dehydrogenase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase, ... | | Authors: | Wischgoll, S, Warkentin, E, Boll, M, Ermler, U. | | Deposit date: | 2010-04-27 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for promoting and preventing decarboxylation in glutaryl-coenzyme a dehydrogenases.
Biochemistry, 49, 2010
|
|
6JTD
 
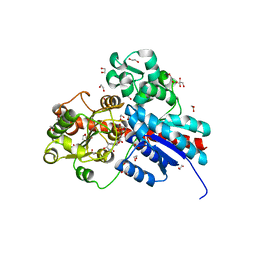 | | Crystal structure of TcCGT1 in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, C-glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Zhao, P, Yun, C.H. | | Deposit date: | 2019-04-10 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular and Structural Characterization of a Promiscuous C-Glycosyltransferase from Trollius chinensis.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
2VNC
 
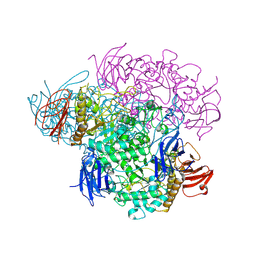 | | Crystal structure of Glycogen Debranching enzyme TreX from Sulfolobus solfataricus | | Descriptor: | GLYCOGEN OPERON PROTEIN GLGX | | Authors: | Song, H.-N, Yoon, S.-M, Cha, H, Park, K.-T, Woo, E.-J. | | Deposit date: | 2008-02-04 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insight Into the Bifunctional Mechanism of the Glycogen-Debranching Enzyme Trex from the Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 283, 2008
|
|
2VDX
 
 | |
1W3U
 
 | |
2XGD
 
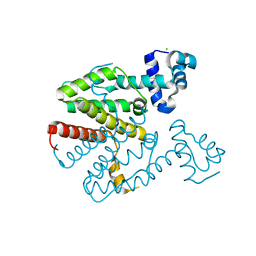 | | Crystal structure of a designed homodimeric variant T-A(L)A(L) of the tetracycline repressor | | Descriptor: | CHLORIDE ION, TETRACYCLINE REPRESSOR PROTEIN CLASS B FROM TRANSPOSON TN10, TETRACYCLINE REPRESSOR PROTEIN CLASS D | | Authors: | Stiebritz, M.T, Wengrzik, S, Richter, J.P, Muller, Y.A. | | Deposit date: | 2010-06-03 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Computational Design of a Chain-Specific Tetracycline Repressor Heterodimer.
J.Mol.Biol., 403, 2010
|
|
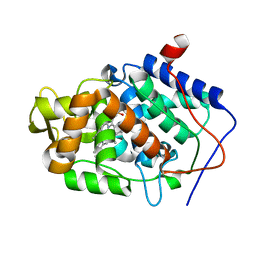
3M25
 
 | |
2XN1
 
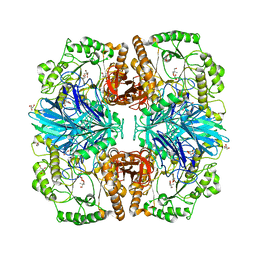 | | Structure of alpha-galactosidase from Lactobacillus acidophilus NCFM with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ALPHA-GALACTOSIDASE, GLYCEROL | | Authors: | Fredslund, F, Abou Hachem, M, Larsen, R.J, Sorensen, P.G, Lo Leggio, L, Svensson, B. | | Deposit date: | 2010-07-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Alpha-Galactosidase from Lactobacillus Acidophilus Ncfm: Insight Into Tetramer Formation and Substrate Binding.
J.Mol.Biol., 412, 2011
|
|
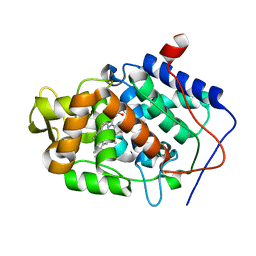
3M2F
 
 | |
3M2W
 
 | | Crystal structure of MAPKAK kinase 2 (MK2) complexed with a spiroazetidine-tetracyclic ATP site inhibitor | | Descriptor: | 2'-(2-fluorophenyl)-1-methyl-6',8',9',11'-tetrahydrospiro[azetidine-3,10'-pyrido[3',4':4,5]pyrrolo[2,3-f]isoquinolin]-7'(5'H)-one, MAGNESIUM ION, MAP kinase-activated protein kinase 2 | | Authors: | Kroemer, M, Revesz, L, Be, C, Izaac, A, Huppertz, C, Schlapbach, A, Scheufler, C. | | Deposit date: | 2010-03-08 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | In vivo and in vitro SAR of tetracyclic MAPKAP-K2 (MK2) inhibitors. Part II.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2V7Z
 
 | | Crystal structure of the 70-kDa heat shock cognate protein from Rattus norvegicus in post-ATP hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK COGNATE 71 KDA PROTEIN, PHOSPHATE ION | | Authors: | Chang, Y.-W, Sun, Y.-J, Wang, C, Hsiao, C.-D. | | Deposit date: | 2007-08-02 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structures of the 70-kDa Heat Shock Proteins in Domain Disjoining Conformation.
J.Biol.Chem., 283, 2008
|
|
