7T2R
 
 | |
6YES
 
 | |
7TBH
 
 | | cryo-EM structure of MBP-KIX-apoferritin complex with peptide 7 | | Descriptor: | Isoform 2 of CREB-binding protein,Ferritin heavy chain, N-terminally processed, LEU-SER-ARG-ARG-PRO-SEP-TYR-ARG-LYS-ILE-LEU-ASN-ASP-LEU-SER-SER-ASP-ALA-PRO | | Authors: | Zhang, K, Horikoshi, N, Li, S, Powers, A, Hameedi, M, Pintilie, G, Chae, H, Khan, Y, Suomivuori, C, Dror, R, Sakamoto, K, Chiu, W, Wakatsuki, S. | | Deposit date: | 2021-12-22 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM, Protein Engineering, and Simulation Enable the Development of Peptide Therapeutics against Acute Myeloid Leukemia.
Acs Cent.Sci., 8, 2022
|
|
7SN7
 
 | |
7N6F
 
 | | Co-complex CYP46A1 with compound 3f | | Descriptor: | (3-fluoroazetidin-1-yl){1-[4-(4-fluorophenyl)pyrimidin-5-yl]piperidin-4-yl}methanone, 1,2-ETHANEDIOL, Cholesterol 24-hydroxylase, ... | | Authors: | Lane, W, Gay, S.C. | | Deposit date: | 2021-06-08 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design and synthesis of aryl-piperidine derivatives as potent and selective PET tracers for cholesterol 24-hydroxylase (CH24H)
Eur.J.Med.Chem., 240, 2022
|
|
6YFM
 
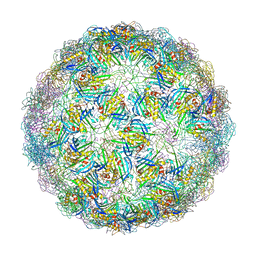 | | Virus-like particle of bacteriophage ESE021 | | Descriptor: | ZINC ION, coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.762 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
7NOZ
 
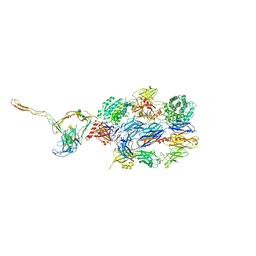 | | Structure of the nanobody stablized properdin bound alternative pathway proconvertase C3b:FB:FP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3 alpha chain, Complement C3 beta chain, ... | | Authors: | Lorenzen, J, Pedersen, D.V, Andersen, G.R. | | Deposit date: | 2021-02-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure determination of an unstable macromolecular complex enabled by nanobody-peptide bridging.
Protein Sci., 31, 2022
|
|
7SP1
 
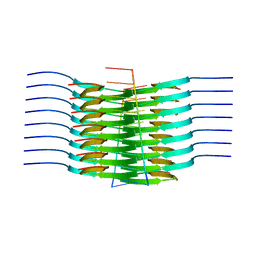 | | RNA-induced tau amyloid fibril | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau, RNA (5'-R(*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3') | | Authors: | Abskharon, R, Sawaya, M.R, Boyer, D.R, Eisenberg, D.S. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of RNA-induced tau fibrils reveals a small C-terminal core that may nucleate fibril formation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6YLF
 
 | |
7N8G
 
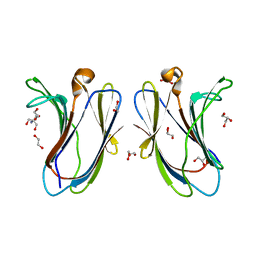 | | Crystal structure of R20A-R22A human Galectin-7 mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2021-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of R20A human Galectin-7 mutant
To Be Published
|
|
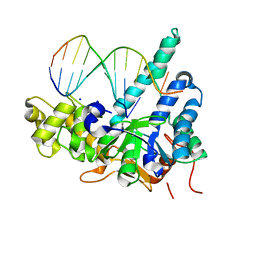
7MXU
 
 | |
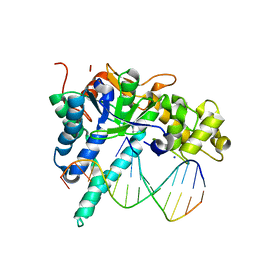
7MXT
 
 | |
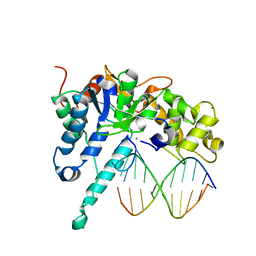
7MXX
 
 | |
7T93
 
 | |
7O1Q
 
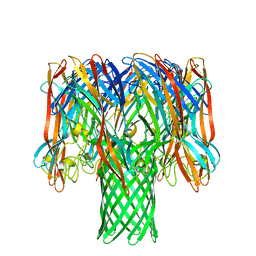 | | Amyloid beta oligomer displayed on the alpha hemolysin scaffold | | Descriptor: | Alpha-hemolysin hybridized Abeta | | Authors: | Wu, J, Blum, T.B, Farrell, D.P, DiMaio, F, Abrahams, J.P, Luo, J. | | Deposit date: | 2021-03-30 | | Release date: | 2021-04-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-electron Microscopy Imaging of Alzheimer's Amyloid-beta 42 Oligomer Displayed on a Functionally and Structurally Relevant Scaffold.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7OD0
 
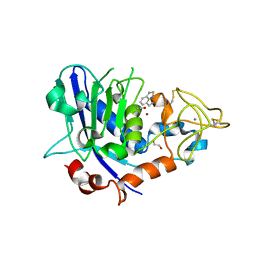 | | Mirolysin in complex with compound 9 | | Descriptor: | 1,2-ETHANEDIOL, 2,1,3-benzothiadiazol-4-ylmethanamine, ACETATE ION, ... | | Authors: | Zak, K.M, Bostock, M.J, Ksiazek, M. | | Deposit date: | 2021-04-28 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Latency, thermal stability, and identification of an inhibitory compound of mirolysin, a secretory protease of the human periodontopathogen Tannerella forsythia .
J Enzyme Inhib Med Chem, 36, 2021
|
|
7SV2
 
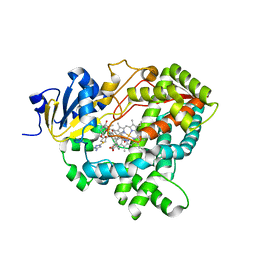 | | Human Cytochrome P450 (CYP) 3A5 ternary complex with azamulin | | Descriptor: | (3aS,4R,5S,6R,8R,9R,9aR,10R)-6-ethyl-5-hydroxy-4,6,9,10-tetramethyl-1-oxodecahydro-3a,9-propanocyclopenta[8]annulen-8-yl [(5-amino-1H-1,2,4-triazol-3-yl)sulfanyl]acetate, Cytochrome P450 3A5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hsu, M, Johnson, E.F. | | Deposit date: | 2021-11-18 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural characterization of the homotropic cooperative binding of azamulin to human cytochrome P450 3A5.
J.Biol.Chem., 298, 2022
|
|
7NZ1
 
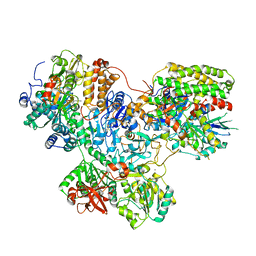 | |
6YHH
 
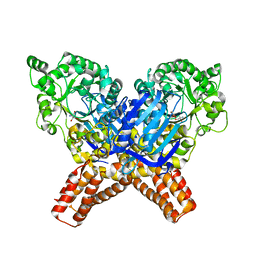 | | X-ray Structure of Flavobacterium johnsoniae chitobiase (FjGH20) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-N-acetylglucosaminidase-like protein Glycoside hydrolase family 20, GLYCEROL | | Authors: | Mazurkewich, S, Helland, R, MacKenzie, A, Eijsink, V.G.H, Pope, P.B, Branden, G, Larsbrink, J. | | Deposit date: | 2020-03-30 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights of the enzymes from the chitin utilization locus of Flavobacterium johnsoniae.
Sci Rep, 10, 2020
|
|
7NYR
 
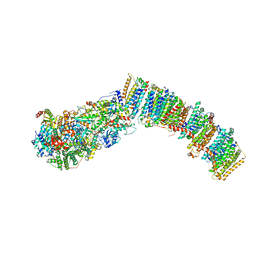 | |
7OAY
 
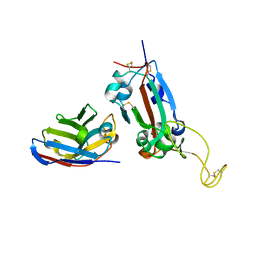 | | Nanobody F2 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2 nanobody, Spike protein S1 | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
2JVW
 
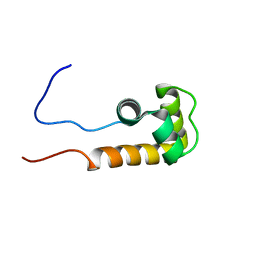 | | Solution NMR structure of uncharacterized protein Q5E7H1 from Vibrio fischeri. Northeast Structural Genomics target VfR117 | | Descriptor: | Uncharacterized protein | | Authors: | Aramini, J.M, Rossi, P, Wang, D, Nwosu, C, Owens, L.A, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-26 | | Release date: | 2007-10-16 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of VF0530 from Vibrio fischeri reveals a nucleic acid-binding function.
Proteins, 79, 2011
|
|
2JP9
 
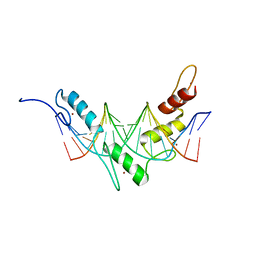 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DGP*DCP*DGP*DC)-3'), DNA (5'-D(P*DGP*DCP*DGP*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-04-30 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
7NYU
 
 | |
7NYV
 
 | |
