2LFU
 
 | | The structure of a N. meningitides protein targeted for vaccine development | | Descriptor: | Gna2132 | | Authors: | Esposito, V, Musi, V, De Chiara, C, Kelly, G, Veggi, D, Pizza, M, Pastore, A. | | Deposit date: | 2011-07-15 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal Domain of Neisseria Heparin Binding Antigen (NHBA), One of the Main Antigens of a Novel Vaccine against Neisseria meningitidis.
J.Biol.Chem., 286, 2011
|
|
6DGC
 
 | | Crystal structure of the C-terminal catalytic domain of ISC1926 TnpA, an IS607-like serine recombinase | | Descriptor: | ISC1926 TnpA C-terminal catalytic domain | | Authors: | Hancock, S.P, Kumar, P, Cascio, D, Johnson, R.C. | | Deposit date: | 2018-05-17 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Multiple serine transposase dimers assemble the transposon-end synaptic complex during IS607-family transposition.
Elife, 7, 2018
|
|
4N4C
 
 | | Crystal structure of the C-terminal swapped dimer of a Bovine seminal ribonuclease mutant | | Descriptor: | PHOSPHATE ION, Seminal ribonuclease | | Authors: | Pica, A, Russo Krauss, I, Merlino, A, Sica, F. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The multiple forms of bovine seminal ribonuclease: Structure and stability of a C-terminal swapped dimer.
Febs Lett., 587, 2013
|
|
3Q9D
 
 | |
2QSA
 
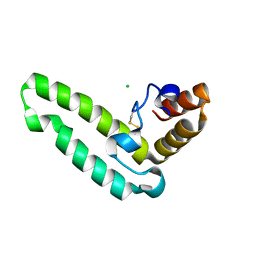 | | Crystal structure of J-domain of DnaJ homolog dnj-2 precursor from C.elegans. | | Descriptor: | CHLORIDE ION, DnaJ homolog dnj-2 | | Authors: | Osipiuk, J, Mulligan, R, Gu, M, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-30 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray crystal structure of J-domain of DnaJ homolog dnj-2 precursor from C.elegans.
To be Published
|
|
5NMS
 
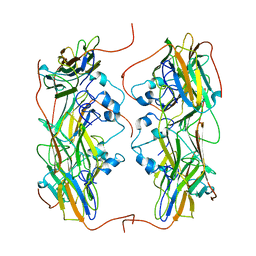 | | Hsp21 dodecamer, structural model based on cryo-EM and homology modelling | | Descriptor: | 25.3 kDa heat shock protein, chloroplastic | | Authors: | Rutsdottir, G, Harmark, J, Koeck, P.J.B, Hebert, H, Soderberg, C.A.G, Emanuelsson, C. | | Deposit date: | 2017-04-07 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural model of dodecameric heat-shock protein Hsp21: Flexible N-terminal arms interact with client proteins while C-terminal tails maintain the dodecamer and chaperone activity.
J. Biol. Chem., 292, 2017
|
|
3LGQ
 
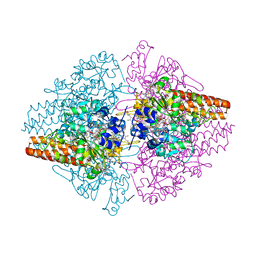 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase in complex with sulfite (modified Tyr-303) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Filimonenkov, A.A, Tikhonova, T.V, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2010-01-21 | | Release date: | 2011-02-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8SKB
 
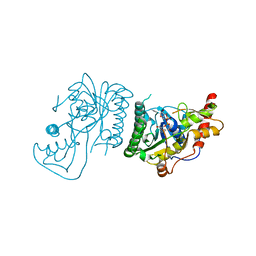 | | Crystal Structure of GDP-mannose 3,5 epimerase de Myrciaria dubia in complex with NAD | | Descriptor: | GDP-mannose 3,5 epimerase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2023-04-19 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit camu-camu.
J.Exp.Bot., 75, 2024
|
|
4JAI
 
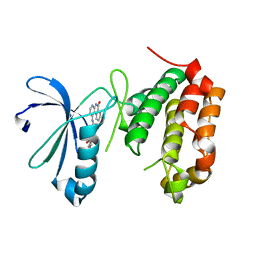 | | Crystal Structure of Aurora Kinase A in complex with N-{4-[(6-oxo-5,6-dihydrobenzo[c][1,8]naphthyridin-1-yl)amino]phenyl}benzamide | | Descriptor: | Aurora kinase A, N-{4-[(6-oxo-5,6-dihydrobenzo[c][1,8]naphthyridin-1-yl)amino]phenyl}benzamide | | Authors: | Jiang, X, Josephson, K, Huck, B, Goutopoulos, A, Karra, S. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | SAR and evaluation of novel 5H-benzo[c][1,8]naphthyridin-6-one analogs as Aurora kinase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3LG1
 
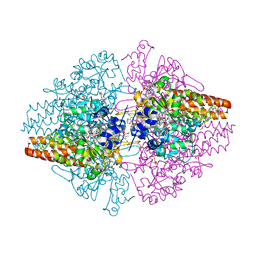 | | Structure of the Thioalkalivibrio nitratireducens cytochrome c nitrite reductase reduced by sodium borohydride (in complex with sulfite) | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CALCIUM ION, Eight-heme nitrite reductase, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Boyko, K.M, Filimonenkov, A.A, Dorovatovsky, P.V, Tikhonova, T.V, Popov, V.O. | | Deposit date: | 2010-01-19 | | Release date: | 2010-12-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Covalent modifications of the catalytic tyrosine in octahaem cytochrome c nitrite reductase and their effect on the enzyme activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5VGE
 
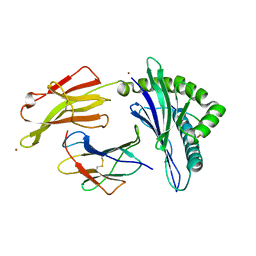 | | Crystal structure of HLA-C*07:02 in complex with RYR peptide | | Descriptor: | ARG-TYR-ARG-PRO-GLY-THR-VAL-ALA-LEU, Beta-2-microglobulin, ZINC ION, ... | | Authors: | Mobbs, J.I, Vivian, J.P, Gras, S, Rossjohn, J. | | Deposit date: | 2017-04-11 | | Release date: | 2017-06-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and regulatory diversity shape HLA-C protein expression levels.
Nat Commun, 8, 2017
|
|
2WJN
 
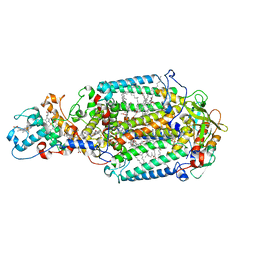 | | Lipidic sponge phase crystal structure of photosynthetic reaction centre from Blastochloris viridis (high dose) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Wohri, A.B, Wahlgren, W.Y, Malmerberg, E, Johansson, L.C, Neutze, R, Katona, G. | | Deposit date: | 2009-05-27 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Lipidic sponge phase crystal structure of a photosynthetic reaction center reveals lipids on the protein surface.
Biochemistry, 48, 2009
|
|
1A56
 
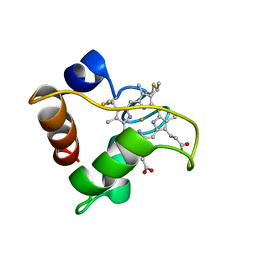 | | PRIMARY SEQUENCE AND SOLUTION CONFORMATION OF FERRICYTOCHROME C-552 FROM NITROSOMONAS EUROPAEA, NMR, MEAN STRUCTURE REFINED WITH EXPLICIT HYDROGEN BOND CONSTRAINTS | | Descriptor: | FERRICYTOCHROME C-552, HEME C | | Authors: | Timkovich, R, Bergmann, D, Arciero, D.M, Hooper, A.B. | | Deposit date: | 1998-02-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Primary sequence and solution conformation of ferrocytochrome c-552 from Nitrosomonas europaea.
Biophys.J., 75, 1998
|
|
4JAJ
 
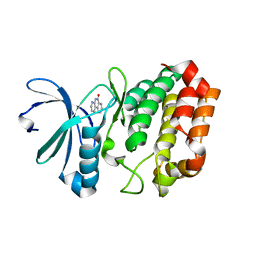 | | Crystal Structure of Aurora Kinase A in complex with BENZO[C][1,8]NAPHTHYRIDIN-6(5H)-ONE | | Descriptor: | Aurora kinase A, benzo[c][1,8]naphthyridin-6(5H)-one | | Authors: | Jiang, X, Josephson, K, Huck, B, Goutopoulos, A, Karra, S. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SAR and evaluation of novel 5H-benzo[c][1,8]naphthyridin-6-one analogs as Aurora kinase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1YNC
 
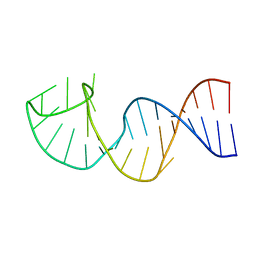 | | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor | | Descriptor: | apolipoprotein B mRNA | | Authors: | Maris, C, Masse, J, Allain, F.H, Chester, A, Navaratnam, N. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor.
Rna, 11, 2005
|
|
1A8C
 
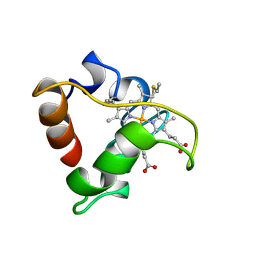 | | PRIMARY SEQUENCE AND SOLUTION CONFORMATION OF FERROCYTOCHROME C-552 FROM NITROSOMONAS EUROPAEA, NMR, MEAN STRUCTURE REFINED WITHOUT HYDROGEN BOND CONSTRAINTS | | Descriptor: | FERROCYTOCHROME C-552, HEME C | | Authors: | Timkovich, R, Bergmann, D, Arciero, D.M, Hooper, A.B. | | Deposit date: | 1998-03-23 | | Release date: | 1998-10-21 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Primary sequence and solution conformation of ferrocytochrome c-552 from Nitrosomonas europaea.
Biophys.J., 75, 1998
|
|
5WSP
 
 | | Crystal structure of DNA3 duplex | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*GP*TP*CP*C)-3'), STRONTIUM ION | | Authors: | Gan, J.H, Liu, H.H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Flexibility and stabilization of HgII-mediated C:T and T:T base pairs in DNA duplex
Nucleic Acids Res., 45, 2017
|
|
5WSR
 
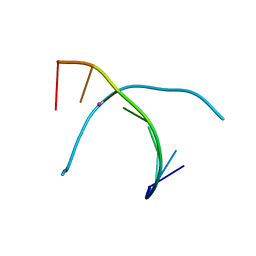 | | Crystal structure of T-Hg-T pair containing DNA duplex | | Descriptor: | DNA (5'-D(*GP*GP*TP*CP*GP*TP*CP*C)-3'), MERCURY (II) ION | | Authors: | Gan, J.H, Liu, H.H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Flexibility and stabilization of HgII-mediated C:T and T:T base pairs in DNA duplex
Nucleic Acids Res., 45, 2017
|
|
3HKY
 
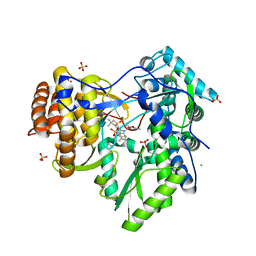 | | HCV NS5B polymerase genotype 1b in complex with 1,5 benzodiazepine 6 | | Descriptor: | (11S)-10-[(2,5-dimethyl-1,3-oxazol-4-yl)carbonyl]-11-{2-fluoro-4-[(2-methylprop-2-en-1-yl)oxy]phenyl}-3,3-dimethyl-2,3,4,5,10,11-hexahydrothiopyrano[3,2-b][1,5]benzodiazepin-6-ol 1,1-dioxide, CHLORIDE ION, RNA-directed RNA polymerase, ... | | Authors: | Nyanguile, O, De Bondt, H.L. | | Deposit date: | 2009-05-26 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1a/1b subtype profiling of nonnucleoside polymerase inhibitors of hepatitis C virus
J.Virol., 84, 2010
|
|
3PSF
 
 | |
5AGF
 
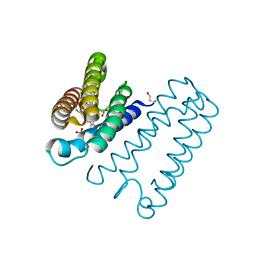 | | Nitrosyl complex of the D121Q variant of cytochrome c prime from Alcaligenes xylosoxidans | | Descriptor: | CYTOCHROME C PRIME, HEME C, NITRIC OXIDE, ... | | Authors: | Gahfoor, D.D, Kekilli, D, Abdullah, G.H, Dworkowski, F.S.N, Hassan, H.G, Wilson, M.T, Hough, M.A, Strange, R.W. | | Deposit date: | 2015-01-30 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Hydrogen Bonding of the Dissociated Histidine Ligand is not Required for Formation of a Proximal No Adduct in Cytochrome C'.
J.Biol.Inorg.Chem., 20, 2015
|
|
5WJ6
 
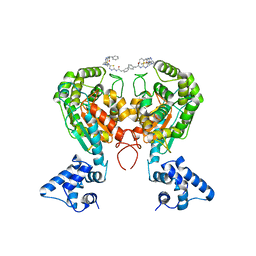 | | Crystal structure of glutaminase C in complex with inhibitor 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}amino)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide (UPGL-00004) | | Descriptor: | 2-phenyl-N-{5-[4-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}amino)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Characterization of the interactions of potent allosteric inhibitors with glutaminase C, a key enzyme in cancer cell glutamine metabolism.
J. Biol. Chem., 293, 2018
|
|
7MQN
 
 | |
5HRX
 
 | | Crystal structure of the fifth bromodomain of human PB1 in complex with 1-butylisochromeno[3,4-c]pyrazol-5(2H)-one) compound | | Descriptor: | 1,2-ETHANEDIOL, 1-butylisochromeno[3,4-c]pyrazol-5(3H)-one, Protein polybromo-1 | | Authors: | Tallant, C, Myrianthopoulos, V, Gaboriaud-Kolar, N, Newman, J.A, Picaud, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Mikros, E, Knapp, S. | | Deposit date: | 2016-01-24 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery and Optimization of a Selective Ligand for the Switch/Sucrose Nonfermenting-Related Bromodomains of Polybromo Protein-1 by the Use of Virtual Screening and Hydration Analysis.
J.Med.Chem., 59, 2016
|
|
9CAI
 
 | | High-resolution C. elegans 80S ribosome structure - class 1 | | Descriptor: | 18S rRNA, 28S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Sehgal, E, Serrao, V.H.B, Arribere, J. | | Deposit date: | 2024-06-17 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | High-resolution reconstruction of a C. elegans ribosome sheds light on evolutionary dynamics and tissue specificity.
Rna, 30, 2024
|
|
