6BGW
 
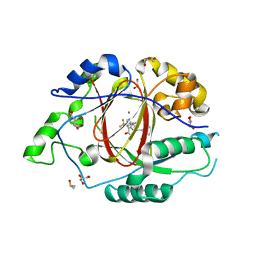 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(4,4-difluoropiperidin-1-yl)ethoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound N41) | | Descriptor: | 2-{(S)-(2-chlorophenyl)[2-(4,4-difluoropiperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.644 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH4
 
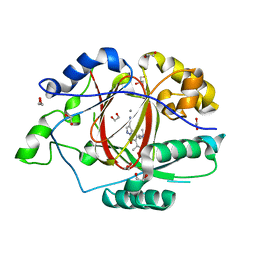 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 5-(1-(tert-butyl)-1H-pyrazol-4-yl)-6-isopropyl-7-oxo-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile (Compound N75/CPI-48) | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-tert-butyl-1H-pyrazol-4-yl)-7-oxo-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH0
 
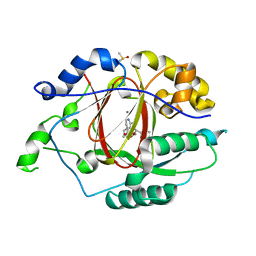 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR (R)-2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1l2-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N51) | | Descriptor: | 2-{(R)-(2-chlorophenyl)[2-(piperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, Lysine-specific demethylase 5A, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BGU
 
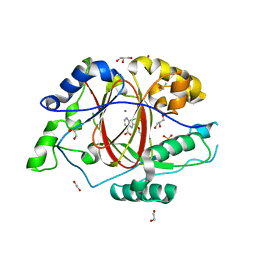 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(propoxy)methyl)-1H-pyrrolo[3,2-b]pyridine (Compound N9) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(R)-(2-chlorophenyl)(propoxy)methyl]-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH2
 
 | |
1SVU
 
 | | Structure of the Q237W mutant of HhaI DNA methyltransferase: an insight into protein-protein interactions | | Descriptor: | Modification methylase HhaI, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Dong, A, Zhou, L, Zhang, X, Stickel, S, Roberts, R.J, Cheng, X. | | Deposit date: | 2004-03-30 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structure of the Q237W mutant of HhaI DNA methyltransferase: an insight into protein-protein interactions
Biol.Chem., 385, 2004
|
|
3CM0
 
 | | Crystal structure of adenylate kinase from Thermus thermophilus HB8 | | Descriptor: | Adenylate kinase | | Authors: | Nakagawa, N, Kondo, N, Masui, R, Matsuda, Z, Iwamoto, I, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-03-20 | | Release date: | 2008-09-23 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of adenylate kinase from Thermus thermophilus HB8.
To be Published
|
|
4TZA
 
 | |
4BWZ
 
 | |
4TZG
 
 | | Crystal structure of eCGP123, an extremely thermostable green fluorescent protein | | Descriptor: | Fluorescent Protein | | Authors: | Close, D.W, Don Paul, C, Traore, D.A.K, Wilce, M.C.J, Prescott, M, Bradbury, A.R.M. | | Deposit date: | 2014-07-10 | | Release date: | 2014-10-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thermal green protein, an extremely stable, nonaggregating fluorescent protein created by structure-guided surface engineering.
Proteins, 83, 2015
|
|
1SKM
 
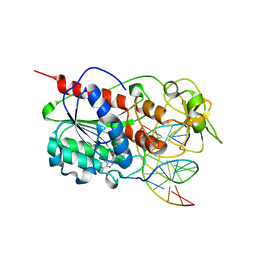 | | HhaI methyltransferase in complex with DNA containing an abasic south carbocyclic sugar at its target site | | Descriptor: | 5'-D(*T*GP*TP*CP*AP*GP*(HCX)P*GP*CP*AP*TP*GP*G)-3', 5'-D(*TP*CP*CP*AP*TP*GP*CP*GP*CP*TP*GP*AP*C)-3', Modification methylase HhaI, ... | | Authors: | Horton, J.R, Ratner, G, Banavali, N, Huang, N, Marquez, V.E, MacKerell, A.D, Cheng, X. | | Deposit date: | 2004-03-05 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caught in the act: visualization of an intermediate in the DNA base-flipping pathway induced by HhaI methyltransferase
Nucleic Acids Res., 32, 2004
|
|
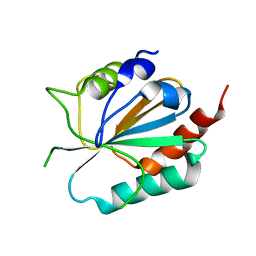
3CMI
 
 | |
4D61
 
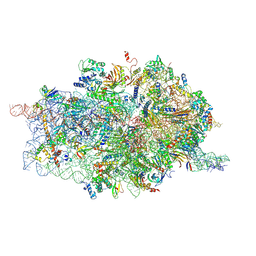 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | 18S RRNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-07 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus Ires.
Mol.Cell, 57, 2015
|
|
4D5L
 
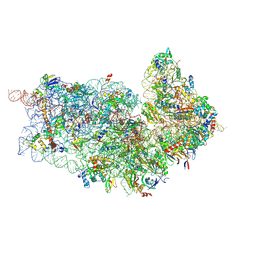 | | Cryo-EM structures of ribosomal 80S complexes with termination factors and cricket paralysis virus IRES reveal the IRES in the translocated state | | Descriptor: | 18S RRNA 2, 40S RIBOSOMAL PROTEIN ES1, 40S RIBOSOMAL PROTEIN ES10, ... | | Authors: | Muhs, M, Hilal, T, Mielke, T, Skabkin, M.A, Sanbonmatsu, K.Y, Pestova, T.V, Spahn, C.M.T. | | Deposit date: | 2014-11-05 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cryo-Em of Ribosomal 80S Complexes with Termination Factors Reveals the Translocated Cricket Paralysis Virus Ires.
Mol.Cell, 57, 2015
|
|
5GJO
 
 | |
5GJN
 
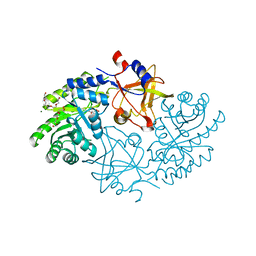 | |
8OZ0
 
 | | Structure of a human 48S translation initiation complex with eIF4F and eIF4A | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Brito Querido, J, Sokabe, M, Diaz-Lopez, I, Gordiyenko, Y, Fraser, C.S, Ramakrishnan, V. | | Deposit date: | 2023-05-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structure of a human translation initiation complex reveals two independent roles for the helicase eIF4A.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7KTR
 
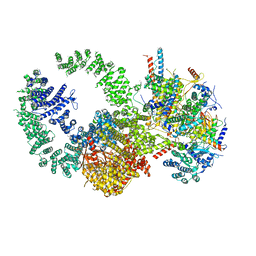 | | Cryo-EM structure of the human SAGA coactivator complex (TRRAP, core) | | Descriptor: | Ataxin-7, INOSITOL HEXAKISPHOSPHATE, Isoform 3 of Transcription factor SPT20 homolog, ... | | Authors: | Herbst, D.A, Esbin, M.N, Nogales, E. | | Deposit date: | 2020-11-24 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structure of the human SAGA coactivator complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
5GJM
 
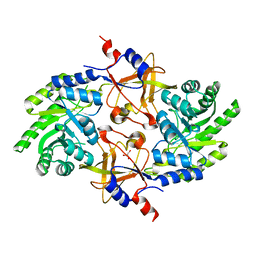 | |
5GJP
 
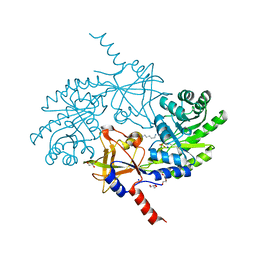 | |
5GW4
 
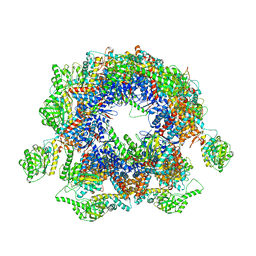 | | Structure of Yeast NPP-TRiC | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Zang, Y, Jin, M, Wang, H, Cong, Y. | | Deposit date: | 2016-09-08 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
5GW5
 
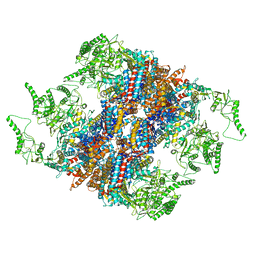 | | Structure of TRiC-AMP-PNP | | Descriptor: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | Authors: | Zang, Y, Jin, M, Wang, H, Cong, Y. | | Deposit date: | 2016-09-08 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
8Q5I
 
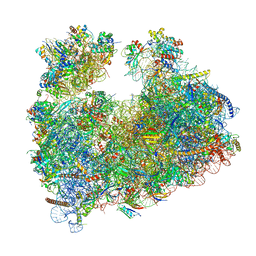 | | Structure of Candida albicans 80S ribosome in complex with cephaeline | | Descriptor: | 18S ribosomal RNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Atamas, A, Guskov, A, Yusupov, M. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural characterization of cephaeline binding to the eukaryotic ribosome using Cryo-Electron Microscopy
Biopolym Cell, 2023
|
|
8PPK
 
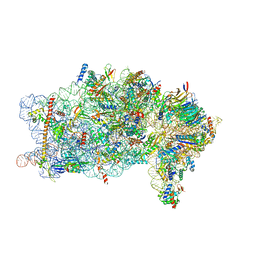 | | Bat-Hp-CoV Nsp1 and eIF1 bound to the human 40S small ribosomal subunit | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
7MHT
 
 | | CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI/DNA COMPLEX | | Descriptor: | 5'-D(P*CP*CP*AP*TP*GP*AP*GP*CP*TP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI, ... | | Authors: | O'Gara, M, Horton, J.R, Roberts, R.J, Cheng, X. | | Deposit date: | 1998-08-05 | | Release date: | 1998-11-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base.
Nat.Struct.Biol., 5, 1998
|
|
