4FNA
 
 | | Structure of unliganded FhuD2 from Staphylococcus Aureus | | Descriptor: | Ferric hydroxamate receptor 2, SULFATE ION | | Authors: | Shilton, B.H, Heinrichs, D.E. | | Deposit date: | 2012-06-19 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal and solution structure analysis of FhuD2 from Staphylococcus aureus in multiple unliganded conformations and bound to ferrioxamine-B.
Biochemistry, 53, 2014
|
|
4F5W
 
 | | Crystal structure of ligand free human STING CTD | | Descriptor: | CALCIUM ION, Transmembrane protein 173 | | Authors: | Gu, L, Shang, G, Zhu, D, Li, N, Zhang, J, Zhu, C, Lu, D, Liu, C, Yu, Q, Zhao, Y, Xu, S. | | Deposit date: | 2012-05-13 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP
Nat.Struct.Mol.Biol., 19, 2012
|
|
3C2S
 
 | | Structural Characterization of a Human Fc Fragment Engineered for Lack of Effector Functions | | Descriptor: | IGHM protein, ZINC ION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Oganesyan, V, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2008-01-25 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of a human Fc fragment engineered for lack of effector functions.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4F65
 
 | | Crystal structure of Human Fibroblast Growth Factor Receptor 1 Kinase domain in complex with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N~2~-[(3-methyl-1,2-oxazol-5-yl)methyl]-N~4~-[3-(2-phenylethyl)-1H-pyrazol-5-yl]pyrimidine-2,4-diamine, Fibroblast growth factor receptor 1, ... | | Authors: | Norman, R.A, Breed, J, Ogg, D. | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Protein-Ligand Crystal Structures Can Guide the Design of Selective Inhibitors of the FGFR Tyrosine Kinase.
J.Med.Chem., 55, 2012
|
|
3V1S
 
 | | Scaffold tailoring by a newly detected Pictet-Spenglerase ac-tivity of strictosidine synthase (STR1): from the common tryp-toline skeleton to the rare piperazino-indole framework | | Descriptor: | 2-(1H-indol-1-yl)ethanamine, Strictosidine synthase | | Authors: | Wu, F, Zhu, H, Sun, L, Rajendran, C, Wang, M, Ren, X, Panjikar, S, Cherkasov, A, Zou, H, Stoeckigt, J. | | Deposit date: | 2011-12-10 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Scaffold Tailoring by a Newly Detected Pictet-Spenglerase Activity of Strictosidine Synthase: From the Common Tryptoline Skeleton to the Rare Piperazino-indole Framework
J.Am.Chem.Soc., 134, 2012
|
|
4FNL
 
 | |
3C82
 
 | | Bacteriophage lysozyme T4 lysozyme mutant K85A/R96H | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Mooers, B.H.M. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
3C32
 
 | |
4F6S
 
 | | Crystal structure of human CDK8/CYCC in complex with compound 7 (1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]urea) | | Descriptor: | 1,2-ETHANEDIOL, 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]urea, Cyclin-C, ... | | Authors: | Schneider, E.V, Boettcher, J, Huber, R, Maskos, K. | | Deposit date: | 2012-05-15 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-kinetic relationship study of CDK8/CycC specific compounds.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4FO0
 
 | | Human actin-related protein Arp8 in its ATP-bound state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 8, CHLORIDE ION, ... | | Authors: | Gerhold, C.B, Lakomek, K, Seifert, F.U, Hopfner, K.-P. | | Deposit date: | 2012-06-20 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Actin-related protein 8 and its contribution to nucleosome binding.
Nucleic Acids Res., 40, 2012
|
|
3C8D
 
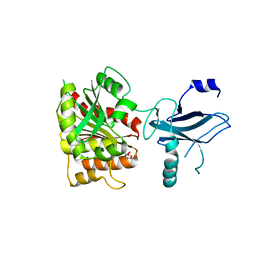 | | Crystal structure of the enterobactin esterase FES from Shigella flexneri in the presence of 2,3-Di-hydroxy-N-benzoyl-glycine | | Descriptor: | CITRIC ACID, Enterochelin esterase | | Authors: | Kim, Y, Maltseva, N, Abergel, R, Holzle, D, Raymond, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-11 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Siderophore Mediated Iron Acquisition: Structure and Specificity of Enterobactin Esterase from Shigella flexneri.
To be Published
|
|
4F7B
 
 | | Structure of the lysosomal domain of limp-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Neculai, D, Ravichandran, M, Seitova, A, Neculai, M, Pizzaro, J.C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Dhe-Paganon, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-05-15 | | Release date: | 2013-10-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36.
Nature, 504, 2013
|
|
3C3F
 
 | |
4F9A
 
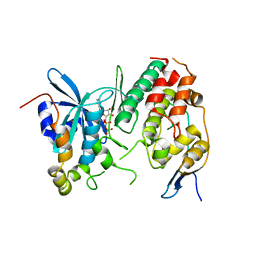 | | Human CDC7 kinase in complex with DBF4 and nucleotide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division cycle 7-related protein kinase, MAGNESIUM ION, ... | | Authors: | Hughes, S, Cherepanov, P. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of human CDC7 kinase in complex with its activator DBF4.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3C3O
 
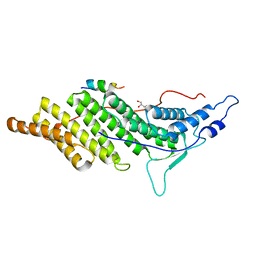 | | ALIX Bro1-domain:CHMIP4A co-crystal structure | | Descriptor: | Charged multivesicular body protein 4a peptide, GLYCEROL, Programmed cell death 6-interacting protein | | Authors: | McCullough, J.B, Fisher, R.D, Whitby, F.G, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-01-28 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | ALIX-CHMP4 interactions in the human ESCRT pathway.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CA4
 
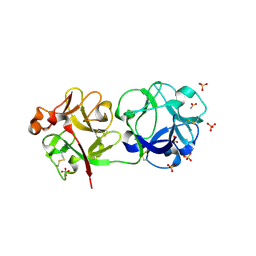 | | Sambucus nigra agglutinin II, tetragonal crystal form- complexed to lactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Maveyraud, L, Mourey, L. | | Deposit date: | 2008-02-19 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for sugar recognition, including the Tn carcinoma antigen, by the lectin SNA-II from Sambucus nigra
Proteins, 75, 2009
|
|
4FP4
 
 | | Crystal structure of isoprenoid synthase a3mx09 (target efi-501993) from pyrobaculum calidifontis | | Descriptor: | GERAN-8-YL GERAN, Polyprenyl synthetase, UNKNOWN ATOM OR ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Poulter, C.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-06-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4FA5
 
 | |
4FB5
 
 | |
3C4B
 
 | | Structure of RNaseIIIb and dsRNA binding domains of mouse Dicer | | Descriptor: | Endoribonuclease Dicer | | Authors: | Lee, J.K, Du, Z, Tjhen, R.J, Stroud, R.M, James, T.L. | | Deposit date: | 2008-01-29 | | Release date: | 2008-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and biochemical insights into the dicing mechanism of mouse Dicer: A conserved lysine is critical for dsRNA cleavage.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CAW
 
 | | Crystal structure of o-succinylbenzoate synthase from Bdellovibrio bacteriovorus liganded with Mg | | Descriptor: | MAGNESIUM ION, o-succinylbenzoate synthase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-20 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4FBD
 
 | | 2.35 Angstrom Crystal Structure of Conserved Hypothetical Protein from Toxoplasma gondii ME49. | | Descriptor: | Putative uncharacterized protein | | Authors: | Minasov, G, Ruan, J, Wawrzak, Z, Shuvalova, L, Ngo, H, Knoll, L, Milligan-Myhre, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom Crystal Structure of Conserved Hypothetical Protein from Toxoplasma gondii ME49.
TO BE PUBLISHED
|
|
3CBC
 
 | | Crystal structure of Siderocalin (NGAL, Lipocalin 2) Y106F complexed with Ferric Enterobactin | | Descriptor: | 2-(2,3-DIHYDROXY-BENZOYLAMINO)-3-HYDROXY-PROPIONIC ACID, GLYCEROL, Neutrophil gelatinase-associated lipocalin, ... | | Authors: | Clifton, M.C, Pizaro, J.C, Strong, R.K. | | Deposit date: | 2008-02-21 | | Release date: | 2008-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The siderocalin/enterobactin interaction: a link between mammalian immunity and bacterial iron transport.
J.Am.Chem.Soc., 130, 2008
|
|
4FPT
 
 | | Carbonic Anhydrase II in complex with ethyl (2Z,4R)-2-(sulfamoylimino)-1,3-thiazolidine-4-carboxylate | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Di Pizio, A, Heine, A, Klebe, G. | | Deposit date: | 2012-06-22 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | High resolution crystal structures of Carbonic Anhydrase II in complex with novel sulfamide binders
To be Published
|
|
4FBL
 
 | | LipS and LipT, two metagenome-derived lipolytic enzymes increase the diversity of known lipase and esterase families | | Descriptor: | CHLORIDE ION, LipS lipolytic enzyme, SPERMIDINE | | Authors: | Chow, J, Krauss, U, Dall Antonia, Y, Fersini, F, Schmeisser, C, Schmidt, M, Menyes, I, Bornscheuer, U, Lauinger, B, Bongen, P, Pietruszka, J, Eckstein, M, Thum, O, Liese, A, Mueller-Dieckmann, J, Jaeger, K.-E, Kovacic, F, Streit, W.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2012-05-23 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Metagenome-Derived Enzymes LipS and LipT Increase the Diversity of Known Lipases.
Plos One, 7, 2012
|
|
