5NC0
 
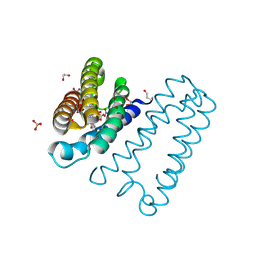 | | The 0.91 A resolution structure of the L16G mutant of cytochrome c prime from Alcaligenes xylosoxidans, complexed with nitric oxide | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome c', HEME C, ... | | Authors: | Strange, R, Hough, M, Antonyuk, S, Rustage, N. | | Deposit date: | 2017-03-02 | | Release date: | 2018-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Distinguishing Nitro vs Nitrito Coordination in Cytochrome c' Using Vibrational Spectroscopy and Density Functional Theory.
Inorg Chem, 56, 2017
|
|
5KKE
 
 | |
6DO3
 
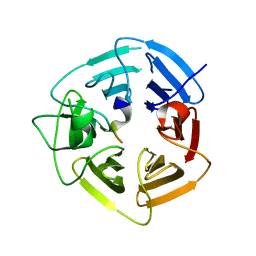 | | KLHDC2 ubiquitin ligase in complex with SelK C-end degron | | Descriptor: | Kelch domain-containing protein 2, SelK C-end Degron | | Authors: | Rusnac, D.V, Lin, H.C, Yen, H.C.S, Zheng, N. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.165 Å) | | Cite: | Recognition of the Diglycine C-End Degron by CRL2KLHDC2Ubiquitin Ligase.
Mol. Cell, 72, 2018
|
|
8EBN
 
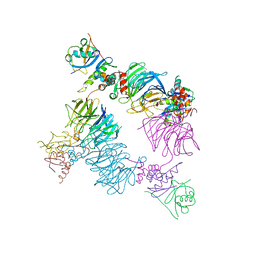 | | Structure of KLHDC2-EloB/C tetrameric assembly | | Descriptor: | Elongin-B, Elongin-C, Kelch domain-containing protein 2 | | Authors: | Scott, D.C, Schulman, B.A. | | Deposit date: | 2022-08-31 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | E3 ligase autoinhibition by C-degron mimicry maintains C-degron substrate fidelity.
Mol.Cell, 83, 2023
|
|
7ZQZ
 
 | |
7ZSW
 
 | |
7ZRX
 
 | |
7ZSV
 
 | |
7ZRW
 
 | |
7ZS4
 
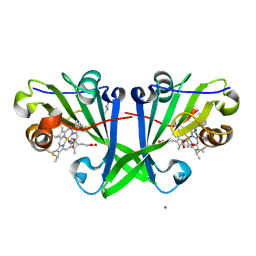 | |
7ZVZ
 
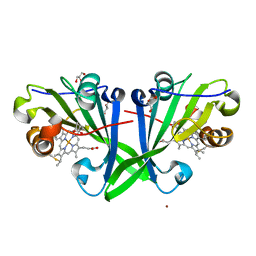 | |
7ZPS
 
 | |
6DO5
 
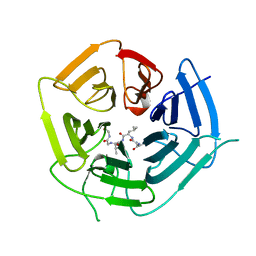 | | KLHDC2 ubiquitin ligase in complex with USP1 C-end degron | | Descriptor: | Kelch domain-containing protein 2, USP1 C-END DEGRON | | Authors: | Rusnac, D.V, Lin, H.C, Yen, H.C.S, Zheng, N. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recognition of the Diglycine C-End Degron by CRL2KLHDC2Ubiquitin Ligase.
Mol. Cell, 72, 2018
|
|
5IP4
 
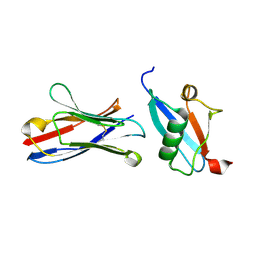 | | X-RAY STRUCTURE OF THE C-TERMINAL DOMAIN OF HUMAN DOUBLECORTIN | | Descriptor: | Neuronal migration protein doublecortin, XA4551 NANOBODY AGAINST C-DCX | | Authors: | Ruf, A, Benz, J, Burger, D, D'Arcy, B, Debulpaep, M, Di Lello, P, Fry, D, Huber, W, Kremer, T, Laeremans, T, Matile, H, Ross, A, Rudolph, M.G, Rufer, A.C, Sharma, A, Steinmetz, M.O, Steyaert, J, Schoch, G, Stihle, M, Thoma, R. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structures of the Human Doublecortin C- and N-terminal Domains in Complex with Specific Antibodies.
J.Biol.Chem., 291, 2016
|
|
6DO4
 
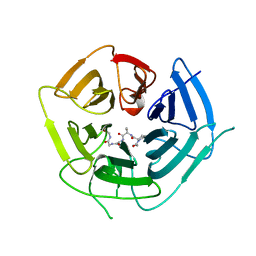 | | KLHDC2 ubiquitin ligase in complex with SelS C-end degron | | Descriptor: | Kelch domain-containing protein 2, SELS C-END DEGRON | | Authors: | Rusnac, D.V, Lin, H.C, Yen, H.C.S, Zheng, N. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of the Diglycine C-End Degron by CRL2KLHDC2Ubiquitin Ligase.
Mol. Cell, 72, 2018
|
|
1AUT
 
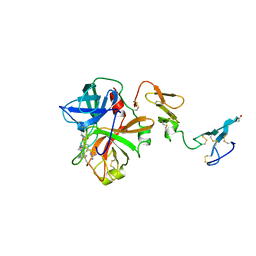 | | Human activated protein C | | Descriptor: | ACTIVATED PROTEIN C, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide | | Authors: | Mather, T, Oganessyan, V, Hof, P, Bode, W, Huber, R, Foundling, S, Esmon, C. | | Deposit date: | 1996-06-08 | | Release date: | 1997-08-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.8 A crystal structure of Gla-domainless activated protein C.
EMBO J., 15, 1996
|
|
1AR1
 
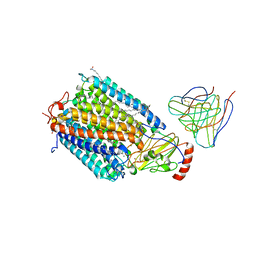 | | Structure at 2.7 Angstrom Resolution of the Paracoccus Denitrificans two-subunit Cytochrome C Oxidase Complexed with an Antibody Fv Fragment | | Descriptor: | ANTIBODY FV FRAGMENT, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Ostermeier, C, Harrenga, A, Ermler, U, Michel, H. | | Deposit date: | 1997-08-08 | | Release date: | 1998-02-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure at 2.7 A resolution of the Paracoccus denitrificans two-subunit cytochrome c oxidase complexed with an antibody FV fragment.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
5KLU
 
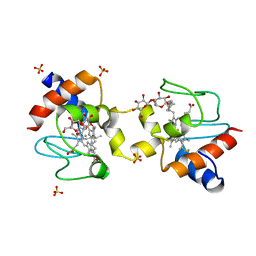 | |
1C2P
 
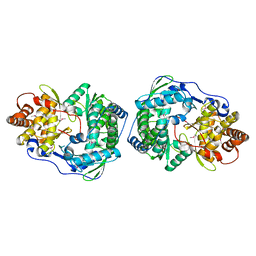 | | HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Descriptor: | RNA-DEPENDENT RNA POLYMERASE | | Authors: | Lesburg, C.A, Cable, M.B, Ferrari, E, Hong, Z, Mannarino, A.F, Weber, P.C. | | Deposit date: | 1999-07-26 | | Release date: | 2000-04-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site.
Nat.Struct.Biol., 6, 1999
|
|
4Q9S
 
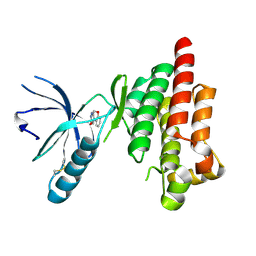 | | Crystal Structure of human Focal Adhesion Kinase (Fak) bound to Compound1 (3,5-DIHYDRO[1,2,4]TRIAZINO[3,4-C][1,4]BENZOXAZIN-2(1H)-ONE) | | Descriptor: | 3,5-dihydro[1,2,4]triazino[3,4-c][1,4]benzoxazin-2(1H)-one, Focal adhesion kinase 1 | | Authors: | Argiriadi, M.A, George, D.M. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of Selective and Orally Bioavailable Protein Kinase C theta (PKC theta ) Inhibitors from a Fragment Hit.
J.Med.Chem., 58, 2015
|
|
5O0Z
 
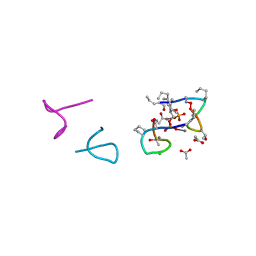 | | Structure of laspartomycin C in complex with geranyl-phosphate | | Descriptor: | ACETIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Vlieg, H.C, Kleijn, L.H.J, Martin, N.I, Janssen, B.J.C. | | Deposit date: | 2017-05-17 | | Release date: | 2017-11-15 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | A High-Resolution Crystal Structure that Reveals Molecular Details of Target Recognition by the Calcium-Dependent Lipopeptide Antibiotic Laspartomycin C.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7VZR
 
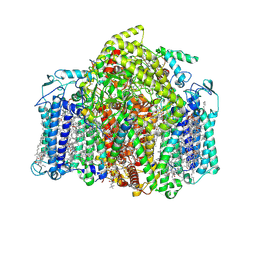 | | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c (the smaller form) | | Descriptor: | BACTERIOCHLOROPHYLL A, CALCIUM ION, CHLOROPHYLL A, ... | | Authors: | Huang, G.Q, Dong, S.S, Qin, X.C, Sui, S.F. | | Deposit date: | 2021-11-16 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.22 Å) | | Cite: | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c
Nat Commun, 13, 2022
|
|
9D1Y
 
 | |
9D8P
 
 | | Focused map of Cryo-EM structure of Ubiquitin C-degron bound to KLHDC10-EloB/C | | Descriptor: | Elongin-B, Elongin-C, Kelch domain-containing protein 10, ... | | Authors: | Chittori, S, Scott, D.C, Schulman, B.A. | | Deposit date: | 2024-08-19 | | Release date: | 2024-11-20 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for C-degron selectivity across KLHDCX family E3 ubiquitin ligases.
Nat Commun, 15, 2024
|
|
9D1Z
 
 | |
