3QKV
 
 | | Crystal structure of fatty acid amide hydrolase with small molecule compound | | Descriptor: | (6-bromo-1'H,4H-spiro[1,3-benzodioxine-2,4'-piperidin]-1'-yl)methanol, Fatty-acid amide hydrolase 1 | | Authors: | Min, X, Walker, N.P.C, Wang, Z. | | Deposit date: | 2011-02-01 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery and molecular basis of potent noncovalent inhibitors of fatty acid amide hydrolase (FAAH).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3QCD
 
 | |
3QLC
 
 | |
3QFF
 
 | | Crystal Structure of ADP complex of purK: N5-carboxyaminoimidazole ribonucleotide synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, N5-carboxyaminoimidazole ribonucleotide synthetase | | Authors: | Fung, L.W, Tuntland, M.L, Santarsiero, B.D, Johnson, M.E. | | Deposit date: | 2011-01-21 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Elucidation of the bicarbonate binding site and insights into the carboxylation mechanism of (N(5))-carboxyaminoimidazole ribonucleotide synthase (PurK) from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3QH3
 
 | | The crystal structure of TCR A6 | | Descriptor: | 1,2-ETHANEDIOL, A6 alpha chain, A6 beta chain, ... | | Authors: | Borbulevych, O.Y, Baker, B.M. | | Deposit date: | 2011-01-25 | | Release date: | 2012-01-04 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Disparate degrees of hypervariable loop flexibility control T-cell receptor cross-reactivity, specificity, and binding mechanism.
J.Mol.Biol., 414, 2011
|
|
3QNR
 
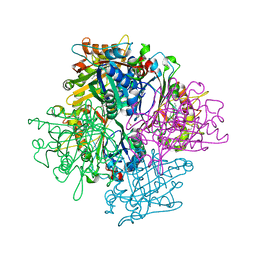 | | DyPB from Rhodococcus jostii RHA1, crystal form 1 | | Descriptor: | DyP Peroxidase, FORMIC ACID, GLYCEROL, ... | | Authors: | Singh, R, Roberts, J.N, Grigg, J.C, Eltis, L.D, Murphy, M.E.P. | | Deposit date: | 2011-02-09 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization of dye-decolorizing peroxidases from Rhodococcus jostii RHA1.
Biochemistry, 50, 2011
|
|
3QI9
 
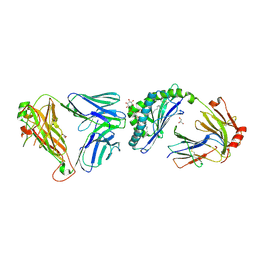 | | Crystal structure of mouse CD1d-alpha-phosphotidylinositol with mouse Valpha14-Vbeta6 2A3-D NKT TCR | | Descriptor: | 2-[(HYDROXY{[(2R,3R,5S,6R)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXYL]OXY}PHOSPHORYL)OXY]-1-[(PALMITOYLOXY)METHYL]ETHYL HEPTADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Clarke, A.J, Rossjohn, J. | | Deposit date: | 2011-01-26 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A molecular basis for NKT cell recognition of CD1d-self-antigen
Immunity, 34, 2011
|
|
3QIW
 
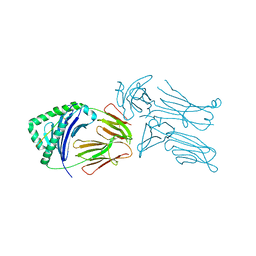 | | Crystal structure of the 226 TCR in complex with MCC-p5E/I-Ek | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H-2 CLASS II HISTOCOMPATIBILITY ANTIGEN, E-K alpha chain, ... | | Authors: | Kruse, A.C, Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
3QJ9
 
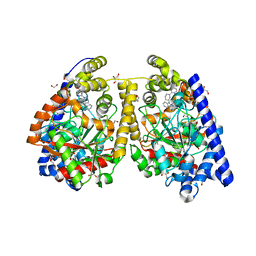 | | Crystal structure of fatty acid amide hydrolase with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-{(3S)-1-[4-(1-benzofuran-2-yl)pyrimidin-2-yl]piperidin-3-yl}-3-ethyl-1,3-dihydro-2H-benzimidazol-2-one, Fatty-acid amide hydrolase 1, ... | | Authors: | Min, X, Walker, N.P.C, Wang, Z. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and molecular basis of potent noncovalent inhibitors of fatty acid amide hydrolase (FAAH).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3QJY
 
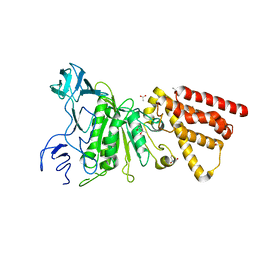 | | Crystal structure of P-loop G234A mutant of subunit A of the A1AO ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Ragunathan, P, Manimekalai, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2011-01-31 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Conserved glycine residues in the P-loop of ATP synthases form a doorframe for nucleotide entrance.
J.Mol.Biol., 413, 2011
|
|
3Q7S
 
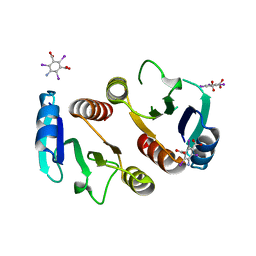 | | 2.1A resolution structure of the ChxR receiver domain containing I3C from Chlamydia trachomatis | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Transcriptional regulatory protein | | Authors: | Hickey, J, Lovell, S, Battaile, K.P, Hu, L, Middaugh, C.R, Hefty, P.S. | | Deposit date: | 2011-01-05 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The atypical response regulator protein ChxR has structural characteristics and dimer interface interactions that are unique within the OmpR/PhoB subfamily.
J.Biol.Chem., 286, 2011
|
|
3Q7V
 
 | |
3QKQ
 
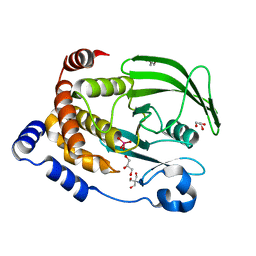 | | Protein Tyrosine Phosphatase 1B - W179F mutant bound with vanadate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1, ... | | Authors: | Brandao, T.A.S, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2011-02-01 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The molecular details of WPD-loop movement differ in the protein-tyrosine phosphatases YopH and PTP1B.
Arch.Biochem.Biophys., 525, 2012
|
|
3Q9C
 
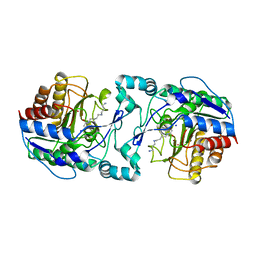 | |
3QLD
 
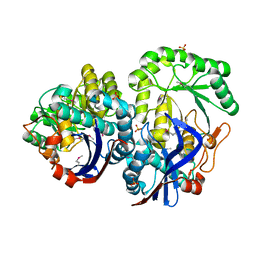 | |
3QB1
 
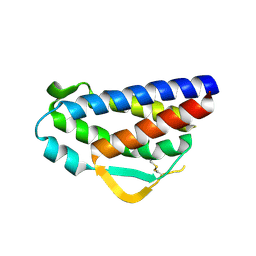 | | Interleukin-2 mutant D10 | | Descriptor: | Interleukin-2 | | Authors: | Levin, A.M, Bates, D.L, Ring, A.M, Lin, J.T, Su, L, Krieg, C, Bowman, G.R, Novick, P, Pande, V.S, Khort, H.E, Boyman, O, Fathman, C.G, Garcia, K.C. | | Deposit date: | 2011-01-12 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Exploiting a natural conformational switch to engineer an interleukin-2 'superkine'
Nature, 484, 2012
|
|
3QNZ
 
 | | Orthorhombic form of IgG1 Fab fragment (in complex with antigenic tubulin peptide) sharing same Fv as IgA | | Descriptor: | Fab fragment of IMMUNOGLOBULIN G1 HEAVY CHAIN, Fab fragment of IMMUNOGLOBULIN G1 LIGHT CHAIN, GLYCEROL, ... | | Authors: | Trajtenberg, F, Correa, A, Buschiazzo, A. | | Deposit date: | 2011-02-09 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a human IgA1 Fab fragment at 1.55 angstrom resolution: potential effect of the constant domains on antigen-affinity modulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3QO6
 
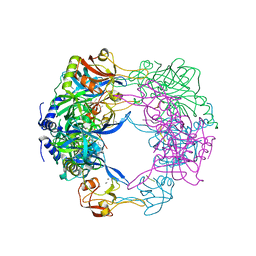 | | Crystal structure analysis of the plant protease Deg1 | | Descriptor: | Protease Do-like 1, chloroplastic, peptide | | Authors: | Clausen, T. | | Deposit date: | 2011-02-09 | | Release date: | 2011-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural adaptation of the plant protease Deg1 to repair photosystem II during light exposure.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QCJ
 
 | |
3QDG
 
 | |
3QPP
 
 | | Structure of PDE10-inhibitor complex | | Descriptor: | 7-methoxy-4-[(3S)-3-phenylpiperidin-1-yl]-6-[2-(quinolin-2-yl)ethoxy]quinazoline, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Pandit, J, Marr, E.S. | | Deposit date: | 2011-02-14 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Use of Structure-Based Design to Discover a Potent, Selective, In Vivo Active Phosphodiesterase 10A Inhibitor Lead Series for the Treatment of Schizophrenia.
J.Med.Chem., 54, 2011
|
|
3QQ8
 
 | |
3Q6U
 
 | | Structure of the apo MET receptor kinase in the dually-phosphorylated, activated state | | Descriptor: | Hepatocyte growth factor receptor | | Authors: | Soisson, S.M, Rickert, K.W, Patel, S.B, Allison, T, Lumb, K.J. | | Deposit date: | 2011-01-03 | | Release date: | 2011-01-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for selective small molecule kinase inhibition of activated c-Met.
J.Biol.Chem., 286, 2011
|
|
3Q8H
 
 | | Crystal structure of 2c-methyl-d-erythritol 2,4-cyclodiphosphate synthase from burkholderia pseudomallei in complex with cytidine derivative EBSI01028 | | Descriptor: | 1,2-ETHANEDIOL, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 5'-deoxy-5'-[(imidazo[2,1-b][1,3]thiazol-5-ylcarbonyl)amino]cytidine, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2011-01-06 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cytidine derivatives as IspF inhibitors of Burkolderia pseudomallei.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3QGD
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid and (2R)-4-(2,6-dimethoxypyrimidin-4-yl)-1-[(4-ethylphenyl)sulfonyl]-N-(4-methoxybenzyl)piperazine-2-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, (2R)-4-(2,6-dimethoxypyrimidin-4-yl)-1-[(4-ethylphenyl)sulfonyl]-N-(4-methoxybenzyl)piperazine-2-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigation of the mode of binding of a novel series of N-benzyl-4-heteroaryl-1-(phenylsulfonyl)piperazine-2-carboxamides to the hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
