4C9S
 
 | | BACTERIAL CHALCONE ISOMERASE IN open CONFORMATION FROM EUBACTERIUM RAMULUS AT 1.8 A RESOLUTION | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2013-10-03 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7ACE
 
 | |
7AFV
 
 | | Crystal structure of tetrameric beta-2-microglobulin deltaN6 S52C stabilized by a covalent ligand | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-oxidanylidene-~{N}-(2-sulfanylethyl)-2,3-dihydro-[1,3]thiazolo[3,2-a]pyrimidine-6-carboxamide, Beta-2-microglobulin | | Authors: | Guthertz, N, Cawood, E, Karamanos, T. | | Deposit date: | 2020-09-20 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Modulation of Amyloidogenic Protein Self-Assembly Using Tethered Small Molecules.
J.Am.Chem.Soc., 142, 2020
|
|
7AFW
 
 | | Beta-Catenin in complex with compound 6 | | Descriptor: | 3-[(2~{R})-4-methyl-5-oxidanylidene-2,3-dihydro-1,4-benzoxazepin-2-yl]benzenecarbonitrile, Catenin beta-1 | | Authors: | Boettcher, J, Kessler, D. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.814 Å) | | Cite: | Getting a Grip on the Undrugged: Targeting beta-Catenin with Fragment-Based Methods.
Chemmedchem, 16, 2021
|
|
6OC4
 
 | | CSP1-cyc(Dab6E10) | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2019-03-21 | | Release date: | 2020-01-08 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity inStreptococcus pneumoniae.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5KPU
 
 | |
6OQ1
 
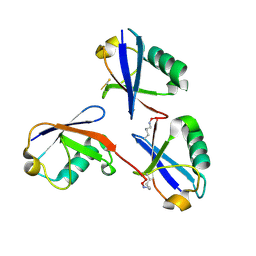 | |
4CBO
 
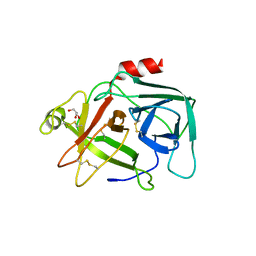 | |
6OPG
 
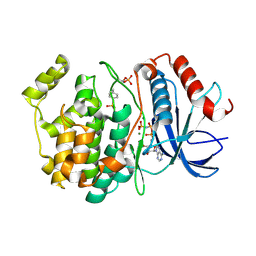 | | phosphorylated ERK2 with AMP-PNP | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Vigers, G.P, Smith, D. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Activation loop dynamics are controlled by conformation-selective inhibitors of ERK2.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7CCO
 
 | | The binding structure of a lanthanide binding tag (LBT3) with lanthanum ion (La3+) | | Descriptor: | LANTHANUM (III) ION, LBT3 | | Authors: | Hatanaka, T, Kikkawa, N, Matsugami, A, Hosokawa, Y, Hayashi, F, Ishida, N. | | Deposit date: | 2020-06-17 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The origins of binding specificity of a lanthanide ion binding peptide.
Sci Rep, 10, 2020
|
|
7CKV
 
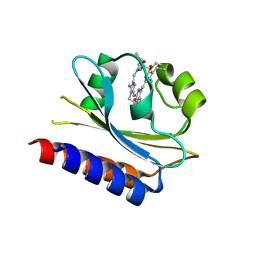 | | Crystal structure of Cyanobacteriochrome GAF domain in Pr state | | Descriptor: | MAGNESIUM ION, PHYCOCYANOBILIN, RcaE | | Authors: | Nagae, T, Koizumi, T, Hirose, Y, Mishima, M. | | Deposit date: | 2020-07-19 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis of the protochromic green/red photocycle of the chromatic acclimation sensor RcaE.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4F2F
 
 | | Crystal structure of the metal binding domain (MBD) of the Streptococcus pneumoniae D39 Cu(I) exporting P-type ATPase CopA with Cu(I) | | Descriptor: | CHLORIDE ION, COPPER (I) ION, Cation-transporting ATPase, ... | | Authors: | Fu, Y, Dann III, C.E, Giedroc, D.P. | | Deposit date: | 2012-05-07 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A new structural paradigm in copper resistance in Streptococcus pneumoniae.
Nat.Chem.Biol., 9, 2013
|
|
4FCR
 
 | | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization | | Descriptor: | 2-{[4-(2-chloro-4,5-dimethoxyphenyl)-5-cyano-7H-pyrrolo[2,3-d]pyrimidin-2-yl]sulfanyl}-N,N-dimethylacetamide, Heat shock protein HSP 90-alpha | | Authors: | Davies, N.G, Browne, H, Davis, B, Foloppe, N, Geoffrey, S, Gibbons, B, Hart, T, Drysdale, M.J, Mansell, H, Massey, A, Matassova, N, Moore, J.D, Murray, J, Pratt, R, Ray, S, Roughley, S.D, Jensen, M.R, Schoepfer, J, Scriven, K, Simmonite, H, Stokes, S, Surgenor, A, Webb, P, Wright, L, Brough, P. | | Deposit date: | 2012-05-25 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization.
Bioorg.Med.Chem., 20, 2012
|
|
5L6K
 
 | | Crystal Structure of Human Carbonic Anhydrase II in Complex with a Quinoline Oligoamide Foldamer | | Descriptor: | 8-azanyl-4-(2-hydroxy-2-oxoethyloxy)quinoline-2-carboxylic acid, 8-azanyl-4-(2-methylpropoxy)quinoline-2-carboxylic acid, 8-azanyl-4-(3-azanylpropoxy)quinoline-2-carboxylic acid, ... | | Authors: | Jewginski, M, Langlois d'Estaintot, B, Granier, T, Huc, Y. | | Deposit date: | 2016-05-30 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Self-Assembled Protein-Aromatic Foldamer Complexes with 2:3 and 2:2:1 Stoichiometries.
J. Am. Chem. Soc., 139, 2017
|
|
5L6U
 
 | | S. ENTERICA HISA MUTANT - D10G, DUP13-15, Q24L, G102A | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase, SULFATE ION | | Authors: | Guo, X, Soderholm, A, Selmer, M. | | Deposit date: | 2016-05-31 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional innovations in the real-time evolution of new ( beta alpha )8 barrel enzymes.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7BXW
 
 | |
4CWB
 
 | | Staphylococcus aureus 7,8-Dihydro-6-hydroxymethylpterin- pyrophosphokinase in complex with AMPCPP and an inhibitor | | Descriptor: | 2-amino-8-[2-oxo-2-(4-phenylphenyl)ethyl]sulfanyl-1,9-dihydropurin-6-one, 7,8-DIHYDRO-6-HYDROXYMETHYLPTERIN-PYROPHOSPHOKINASE, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Dennis, M.L, Swarbrick, J.D, Peat, T.S. | | Deposit date: | 2014-04-02 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure-Based Design and Development of Functionalized Mercaptoguanine Derivatives as Inhibitors of the Folate Biosynthesis Pathway Enzyme 6-Hydroxymethyl-7,8-Dihydropterin Pyrophosphokinase from Staphylococcus Aureus.
J.Med.Chem., 57, 2014
|
|
4CYU
 
 | | Staphylococcus aureus 7,8-Dihydro-6-hydroxymethylpterin- pyrophosphokinase in complex with AMPCPP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 7,8-DIHYDRO-6-HYDROXYMETHYLPTERIN-PYROPHOSPHOKINASE, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Dennis, M.L, Swarbrick, J.D, Peat, T.S. | | Deposit date: | 2014-04-15 | | Release date: | 2015-01-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Based Design and Development of Functionalized Mercaptoguanine Derivatives as Inhibitors of the Folate Biosynthesis Pathway Enzyme 6-Hydroxymethyl-7,8-Dihydropterin Pyrophosphokinase from Staphylococcus Aureus.
J.Med.Chem., 57, 2014
|
|
7BE3
 
 | | Human Galectin-3 in complex with LacdiNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Trovao, F, Carvalho, A.L. | | Deposit date: | 2020-12-22 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Insights into the Molecular Recognition Mechanism of the Cancer and Pathogenic Epitope, LacdiNAc by Immune-Related Lectins.
Chemistry, 27, 2021
|
|
4D06
 
 | | Bacterial chalcone isomerase complexed with naringenin | | Descriptor: | (2E)-3-(4-hydroxyphenyl)-1-(2,4,6-trihydroxyphenyl)prop-2-en-1-one, CHALCONE ISOMERASE, CHLORIDE ION, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-04-24 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4D2G
 
 | | Crystal structure of human PCNA in complex with p15 peptide | | Descriptor: | P15, PROLIFERATING CELL NUCLEAR ANTIGEN | | Authors: | DeBiasio, A, Ibanez, A, Mortuza, G, Molina, R, Cordeiro, T.N, Castillo, F, Villate, M, Merino, N, Lelli, M, Diercks, T, Luque, I, Bernardo, P, Montoya, G, Blanco, F.J. | | Deposit date: | 2014-05-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of P15(Paf)-PCNA Complex and Implications for Clamp Sliding During DNA Replication and Repair.
Nat.Commun., 6, 2015
|
|
6ZPN
 
 | | Crystal structure of Chaetomium thermophilum Raptor | | Descriptor: | WD_REPEATS_REGION domain-containing protein | | Authors: | Imseng, S, Boehm, R, Jakob, R.P, Hall, M.N, Hiller, S, Maier, T. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The dynamic mechanism of 4E-BP1 recognition and phosphorylation by mTORC1.
Mol.Cell, 81, 2021
|
|
7A74
 
 | | Structure of G132N BlaC from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, GLYCEROL, ... | | Authors: | Chikunova, A, Ahmad, M.U, Perrakis, A, Ubbink, M. | | Deposit date: | 2020-08-27 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two beta-Lactamase Variants with Reduced Clavulanic Acid Inhibition Display Different Millisecond Dynamics.
Antimicrob.Agents Chemother., 65, 2021
|
|
4FQO
 
 | | Crystal Structure of Calcium-Loaded S100B Bound to SBi4211 | | Descriptor: | 4,4'-[heptane-1,7-diylbis(oxy)]dibenzenecarboximidamide, CALCIUM ION, Protein S100-B | | Authors: | McKnight, L.E, Raman, E.P, Bezawada, P, Kudrimoti, S, Wilder, P.T, Hartman, K.G, Toth, E.A, Coop, A, MacKerrell, A.D, Weber, D.J. | | Deposit date: | 2012-06-25 | | Release date: | 2012-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Based Discovery of a Novel Pentamidine-Related Inhibitor of the Calcium-Binding Protein S100B.
ACS Med Chem Lett, 3, 2012
|
|
7CG5
 
 | |
