7QCO
 
 | | The structure of Photosystem I tetramer from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Semchonok, D.A, Mondal, J, Cooper, J.C, Schlum, K, Li, M, Amin, M, Sorzano, C.O.S, Ramirez-Aportela, E, Kastritis, P.L, Boekema, E.J, Guskov, A, Bruce, B.D. | | Deposit date: | 2021-11-24 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a tetrameric photosystem I from Chroococcidiopsis TS-821, a thermophilic, unicellular, non-heterocyst-forming cyanobacterium.
Plant Commun., 3, 2022
|
|
6R9S
 
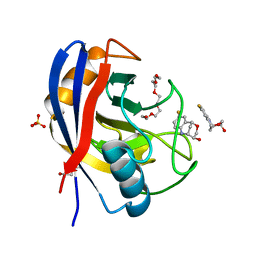 | | Human Cyclophilin D in complex with bicyclic fragment | | Descriptor: | (1~{R},9~{R},10~{S})-4-fluoranyl-12-oxa-8-azatricyclo[7.3.1.0^{2,7}]trideca-2(7),3,5-trien-10-ol, PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Graedler, U. | | Deposit date: | 2019-04-04 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel Cyclophilin D inhibitors starting from three dimensional fragments with millimolar potencies.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6OD8
 
 | |
6OX2
 
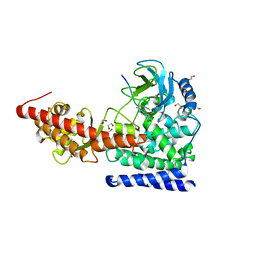 | | SETD3in Complex with an Actin Peptide with the Target Histidine Fully Methylated | | Descriptor: | 1,2-ETHANEDIOL, Actin Peptide, GLYCEROL, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Structural basis for the target specificity of actin histidine methyltransferase SETD3.
Nat Commun, 10, 2019
|
|
4XYB
 
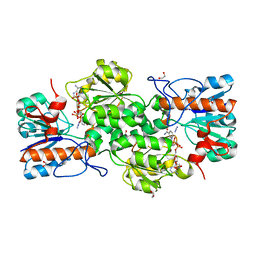 | | GRANULICELLA M. FORMATE DEHYDROGENASE (FDH) IN COMPLEX WITH NADP(+) AND NaN3 | | Descriptor: | 1,2-ETHANEDIOL, AZIDE ION, Formate dehydrogenase, ... | | Authors: | Cendron, L, Fogal, S, Beneventi, E, Bergantino, E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural basis for double cofactor specificity in a new formate dehydrogenase from the acidobacterium Granulicella mallensis MP5ACTX8.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
6S0G
 
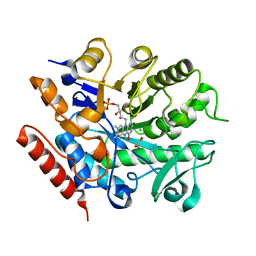 | | Crystal structure of ene-reductase GsOYE from Galdieria sulphuraria | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Robescu, M.R, Niero, M, Hall, M, Bergantino, E, Cendron, L. | | Deposit date: | 2019-06-14 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Two new ene-reductases from photosynthetic extremophiles enlarge the panel of old yellow enzymes: CtOYE and GsOYE.
Appl.Microbiol.Biotechnol., 104, 2020
|
|
6P0B
 
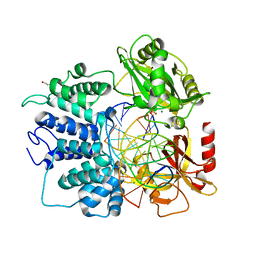 | | Human DNA Ligase 1 (E346A/E592A) Bound to an Adenylated, dideoxy Terminated DNA nick with 200 mM Mg2+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*C)-3'), ... | | Authors: | Schellenberg, M.J, Williams, R.S, Tumbale, P.S, Riccio, A.A. | | Deposit date: | 2019-05-16 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Two-tiered enforcement of high-fidelity DNA ligation.
Nat Commun, 10, 2019
|
|
6VS6
 
 | |
7V3A
 
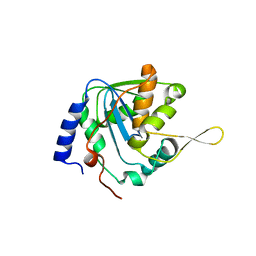 | | Crystal structure of apo-NP exonuclease C409A | | Descriptor: | MAGNESIUM ION, Nucleoprotein, ZINC ION | | Authors: | Hsiao, Y.Y, Huang, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeted Covalent Inhibitors Allosterically Deactivate the DEDDh Lassa Fever Virus NP Exonuclease from Alternative Distal Sites.
Jacs Au, 1, 2021
|
|
7V38
 
 | | Crystal structure of NP exonuclease-PCMPS complex | | Descriptor: | Nucleoprotein, PARA-MERCURY-BENZENESULFONIC ACID, ZINC ION | | Authors: | Hsiao, Y.Y, Huang, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Targeted Covalent Inhibitors Allosterically Deactivate the DEDDh Lassa Fever Virus NP Exonuclease from Alternative Distal Sites.
Jacs Au, 1, 2021
|
|
6P0A
 
 | | Human DNA Ligase 1 Bound to an Adenylated, dideoxy Terminated DNA nick with 2 mM Mg2+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*C)-3'), DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Schellenberg, M.J, Williams, R.S, Tumbale, P.S, Riccio, A.A. | | Deposit date: | 2019-05-16 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Two-tiered enforcement of high-fidelity DNA ligation.
Nat Commun, 10, 2019
|
|
6P0D
 
 | | Human DNA Ligase 1 (E346A/E592A) Bound to an Adenylated, hydroxyl terminated DNA nick | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*C)-3'), ... | | Authors: | Schellenberg, M.J, Williams, R.S, Tumbale, P.S, Riccio, A.A. | | Deposit date: | 2019-05-16 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Two-tiered enforcement of high-fidelity DNA ligation.
Nat Commun, 10, 2019
|
|
6P0C
 
 | | Human DNA Ligase 1 Bound to an Adenylated, hydroxyl terminated DNA nick in EDTA | | Descriptor: | ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*C)-3'), ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Riccio, A.A, Williams, R.S. | | Deposit date: | 2019-05-16 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Two-tiered enforcement of high-fidelity DNA ligation.
Nat Commun, 10, 2019
|
|
6XW5
 
 | | Crystal structure of murine norovirus P domain in complex with Nanobody NB-5820 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Capsid protein, ... | | Authors: | Kilic, T, Sabin, C, Hansman, G. | | Deposit date: | 2020-01-23 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Nanobody-Mediated Neutralization Reveals an Achilles Heel for Norovirus.
J.Virol., 94, 2020
|
|
6P1Z
 
 | |
9L5X
 
 | | Crystal structure of Klebsiella pneumoniae Enoyl-Acyl Carrier Protein Reductase (FabI) in complex with Triclosan | | Descriptor: | 1,2-ETHANEDIOL, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | Authors: | Biswas, S, Patra, A, Kushwaha, G.S, Suar, M. | | Deposit date: | 2024-12-23 | | Release date: | 2025-10-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of Klebsiella pneumoniae Enoyl-Acyl Carrier Protein Reductase (FabI) in complex with Triclosan
To Be Published
|
|
6U9U
 
 | | Structure of GM9_TH8seq732127 FAB | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, GM9_TH8seq732127 FAB heavy chain, ... | | Authors: | Singh, S, Liban, T.J, Pancera, M. | | Deposit date: | 2019-09-09 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Extensive dissemination and intraclonal maturation of HIV Env vaccine-induced B cell responses.
J.Exp.Med., 217, 2020
|
|
8EIW
 
 | | Cobalt(II)-substituted Horse Liver Alcohol Dehydrogenase in Complex with NADH and N-Cyclohexylformamide | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alcohol dehydrogenase E chain, COBALT (II) ION, ... | | Authors: | Zheng, C, Mathews, I.I, Boxer, S.G. | | Deposit date: | 2022-09-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Enhanced active-site electric field accelerates enzyme catalysis.
Nat.Chem., 15, 2023
|
|
8YEQ
 
 | |
4UTZ
 
 | | Crystal structure of zebrafish Sirtuin 5 in complex with adipoylated CPS1-peptide | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CARBAMOYLPHOSPHATE SYNTHETASE I, ... | | Authors: | Pannek, M, Gertz, M, Steegborn, C. | | Deposit date: | 2014-07-24 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Chemical Probing of the Human Sirtuin 5 Active Site Reveals its Substrate Acyl Specificity and Peptide-Based Inhibitors.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8CWL
 
 | | Cryo-EM structure of Human 15-PGDH in complex with small molecule SW222746 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 15-hydroxyprostaglandin dehydrogenase [NAD(+)], 2-methyl-6-[7-(piperidine-1-carbonyl)quinoxalin-2-yl]isoquinolin-1(2H)-one | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-05-19 | | Release date: | 2023-03-08 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Small molecule inhibitors of 15-PGDH exploit a physiologic induced-fit closing system.
Nat Commun, 14, 2023
|
|
6PNV
 
 | | 1.42 Angstrom Resolution Crystal Structure of Translocation Protein TolB from Salmonella enterica | | Descriptor: | POTASSIUM ION, SODIUM ION, Tol-Pal system protein TolB | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-07-03 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | 1.42 Angstrom Resolution Crystal Structure of Translocation Protein TolB from Salmonella enterica
To Be Published
|
|
8SV5
 
 | | Crystal structure of Bacillus anthracis dihydroneopterin aldolase in complex with 6-hydroxymethyl-7,8-dihydropterin | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 7,8-dihydroneopterin aldolase | | Authors: | Shaw, G.X, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2023-05-15 | | Release date: | 2023-06-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of Bacillus anthracis dihydroneopterin aldolase in complex with 6-hydroxymethyl-7,8-dihydropterin
To be published
|
|
8YRL
 
 | | Crystal structure of Aspergillus fumigatus Galactofuranosylransferase (AfGfsA) in complex with UDP and galactofuranose | | Descriptor: | GLYCEROL, Glycosyltransferase family 31 protein, MANGANESE (II) ION, ... | | Authors: | Okuno, A, Oka, T, Chihara, Y, Hirata, R, Kadooka, C, Teramoto, T, Hira, D, Kakuta, Y. | | Deposit date: | 2024-03-21 | | Release date: | 2024-10-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Substrate binding and catalytic mechanism of UDP-alpha-D-galactofuranose: beta-galactofuranoside beta-(1→5)-galactofuranosyltransferase GfsA.
Pnas Nexus, 3, 2024
|
|
6VS9
 
 | |
