7PBN
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s3 [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBQ
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0+A [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBS
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0+A [t1 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
6FUG
 
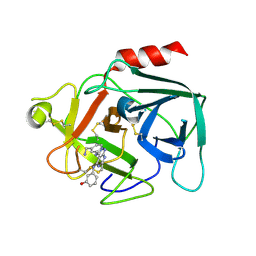 | | Complement factor D in complex with the inhibitor 3-((3-((3-(aminomethyl)phenyl)amino)-1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino)phenol | | Descriptor: | 3-[[3-[[3-(aminomethyl)phenyl]amino]-1~{H}-pyrazolo[3,4-d]pyrimidin-4-yl]amino]phenol, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
7PBR
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0-A [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB, MAGNESIUM ION, ... | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
6FR0
 
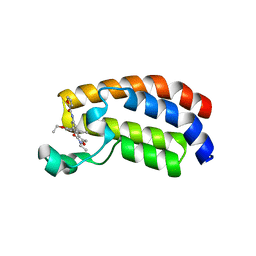 | | Crystal structure of CREBBP bromodomain complexd with PB08 | | Descriptor: | CREB-binding protein, ~{N}-[3-(5-ethanoyl-2-ethoxy-phenyl)-5-(2-ethyl-5-methyl-3-oxidanylidene-1,2-oxazol-4-yl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-15 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
6FRF
 
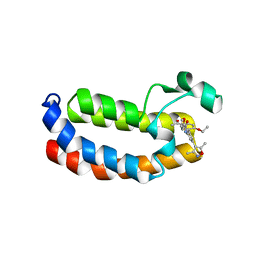 | | Crystal structure of CREBBP bromodomain complexd with PA10 | | Descriptor: | CREB-binding protein, ~{N}-[3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-(5-ethanoyl-2-ethoxy-phenyl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-15 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
8VXD
 
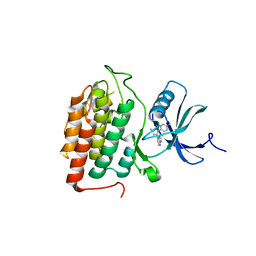 | |
8VXF
 
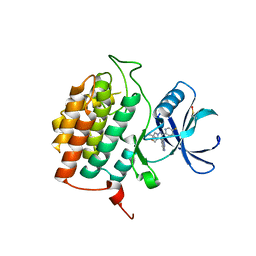 | | Structure of Casein kinase I isoform delta (CK1d) complexed with inhibitor 15 | | Descriptor: | (2P,3P,8S)-2-(5-fluoropyridin-2-yl)-6,6-dimethyl-3-(1H-pyrazolo[3,4-b]pyridin-4-yl)-6,7-dihydro-4H-pyrazolo[5,1-c][1,4]oxazine, Casein kinase I isoform delta | | Authors: | Thompson, A.A, Milligan, C.M, Sharma, S. | | Deposit date: | 2024-02-04 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-Based Optimization of Selective and Brain Penetrant CK1 delta Inhibitors for the Treatment of Circadian Disruptions.
Acs Med.Chem.Lett., 15, 2024
|
|
6FUT
 
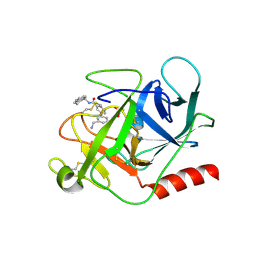 | | Complement factor D in complex with the inhibitor (S)-3'-(aminomethyl)-N-(1,2,3,4-tetrahydronaphthalen-1-yl)-[1,1'-biphenyl]-3-carboxamide | | Descriptor: | 3-[3-(aminomethyl)phenyl]-~{N}-[(1~{S})-1,2,3,4-tetrahydronaphthalen-1-yl]benzamide, Complement factor D, SUCCINIC ACID | | Authors: | Mac Sweeney, A, Vulpetti, A, Erbel, P, Lorthiois, E, Maibaum, J, Randl, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
7BV6
 
 | | Crystal structure of the autophagic STX17/SNAP29/VAMP8 SNARE complex | | Descriptor: | Synaptosomal-associated protein 29, Syntaxin-17, Vesicle-associated membrane protein 8 | | Authors: | Li, Y, Pan, L.F. | | Deposit date: | 2020-04-09 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8VQ2
 
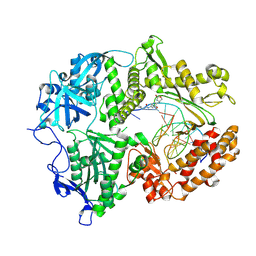 | | HSV1 polymerase ternary complex with dsDNA and compound 44 | | Descriptor: | 2-(4-bromophenyl)-N-(3-methoxy-4-{[(4S)-2-oxo-1,3-oxazolidin-4-yl]methyl}phenyl)acetamide, DNA (5'-D(P*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*TP*AP*C)-3'), DNA (5'-D(P*TP*GP*GP*TP*AP*GP*GP*GP*GP*AP*AP*GP*GP*AP*T)-3'), ... | | Authors: | Hayes, R.P, Heo, M.R, Plotkin, M. | | Deposit date: | 2024-01-17 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (3.829 Å) | | Cite: | Discovery of Broad-Spectrum Herpes Antiviral Oxazolidinone Amide Derivatives and Their Structure-Activity Relationships.
Acs Med.Chem.Lett., 15, 2024
|
|
4TYT
 
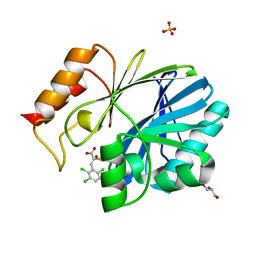 | | Crystal Structure of BcII metallo-beta-lactamase in complex with ML302F | | Descriptor: | (2Z)-2-sulfanyl-3-(2,3,6-trichlorophenyl)prop-2-enoic acid, Beta-lactamase 2, GLYCEROL, ... | | Authors: | Brem, J, van Berkel, S.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2014-07-09 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Rhodanine hydrolysis leads to potent thioenolate mediated metallo-beta-lactamase inhibition.
Nat.Chem., 6, 2014
|
|
8G29
 
 | | Crystal structure of the A2503-C2,C8-dimethylated Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.55A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Wu, K.J.Y, Tresco, B.I.C, Syroegin, E.A, Killeavy, E.E, Balasanyants, S.M, Svetlov, M.S, Gregory, S.T, Atkinson, G.C, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of Cfr-mediated antimicrobial resistance and mechanisms to evade it.
Nat.Chem.Biol., 20, 2024
|
|
6Q55
 
 | | Crystal structure of Cryptosporidium hominis CPSF3 in complex with Compound 61 | | Descriptor: | 3-[7,7-bis(oxidanyl)-8-oxa-7-boranuidabicyclo[4.3.0]nona-1(6),2,4-trien-5-yl]propanoic acid, Cleavage and Polyadenylation Specificity Factor 3 (CPSF3), GLYCEROL, ... | | Authors: | Palencia, A, Swale, C. | | Deposit date: | 2018-12-07 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal-captured inhibition of pre-mRNA processing activity by CPSF3 controls Cryptosporidium infection.
Sci Transl Med, 11, 2019
|
|
5H8G
 
 | | Crystal structure of CK2 with compound 7b | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and Selective CK2 Kinase Inhibitors with Effects on Wnt Pathway Signaling in Vivo.
Acs Med.Chem.Lett., 7, 2016
|
|
7EZR
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-ethyl-7-nitro-3-[3-oxidanylidene-3-(thiophen-2-ylsulfonylamino)propyl]-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZP
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-(3-hydroxy-3-oxopropyl)-5-(2-methylpropyl)-7-nitro-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZF
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 7-chloranyl-5-ethyl-3-(3-hydroxy-3-oxopropyl)-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
5I8B
 
 | | CBP in complex with Cpd23 ((R)-6-(3-(benzyloxy)phenyl)-4-methyl-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one) | | Descriptor: | (4R)-6-[3-(benzyloxy)phenyl]-4-methyl-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5218 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
5H8B
 
 | | Crystal structure of CK2 with compound 2 | | Descriptor: | 1,2-ETHANEDIOL, Casein kinase II subunit alpha, SULFATE ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Potent and Selective CK2 Kinase Inhibitors with Effects on Wnt Pathway Signaling in Vivo.
Acs Med.Chem.Lett., 7, 2016
|
|
5IVQ
 
 | |
6FTY
 
 | | COMPLEMENT FACTOR D COMPLEXED WITH COMPOUND 5 | | Descriptor: | 4-[[(5~{S},7~{R})-3-azanyl-1-adamantyl]carbonylamino]-1~{H}-indole-2-carboxamide, Complement factor D, GLYCEROL | | Authors: | Ostermann, N. | | Deposit date: | 2018-02-26 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
6FUJ
 
 | | Complement factor D in complex with the inhibitor N-(3'-(aminomethyl)-[1,1'-biphenyl]-3-yl)-3-methylbutanamide | | Descriptor: | Complement factor D, ~{N}-[3-[3-(aminomethyl)phenyl]phenyl]-3-methyl-butanamide | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
5I8G
 
 | | CBP in complex with Cpd637 ((R)-4-methyl-6-(1-methyl-3-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-5-yl)-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one) | | Descriptor: | (4R)-4-methyl-6-[1-methyl-3-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-5-yl]-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
