4R9R
 
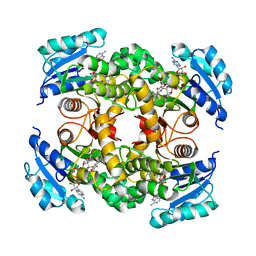 | | Mycobacterium tuberculosis InhA bound to NITD-564 | | Descriptor: | 6-(cyclohexylmethyl)-4-hydroxy-3-phenylpyridin-2(1H)-one, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Noble, C.G. | | Deposit date: | 2014-09-07 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Direct inhibitors of InhA are active against Mycobacterium tuberculosis
Sci Transl Med, 7, 2015
|
|
7U5B
 
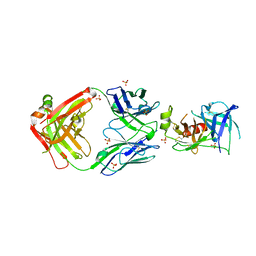 | | Structure of Human KLK5 bound to anti-KLK5 Fab | | Descriptor: | Kallikrein-5, SULFATE ION, anti-KLK5 Fab Heavy Chain, ... | | Authors: | Yin, J, Sudhamsu, J. | | Deposit date: | 2022-03-02 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.371 Å) | | Cite: | Dual antibody inhibition of KLK5 and KLK7 for Netherton syndrome and atopic dermatitis.
Sci Transl Med, 14, 2022
|
|
5C42
 
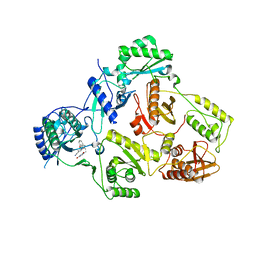 | | Crystal Structure of HIV-1 Reverse Transcriptase (K101P) Variant in Complex with 8-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)indolizine-2-carbonitrile (JLJ555), a non-nucleoside inhibitor | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}indolizine-2-carbonitrile, HIV-1 Reverse Transcriptase, p51 subunit, ... | | Authors: | Frey, K.M, Gray, W.T, Anderson, K.S. | | Deposit date: | 2015-06-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Potent Inhibitors Active against HIV Reverse Transcriptase with K101P, a Mutation Conferring Rilpivirine Resistance.
Acs Med.Chem.Lett., 6, 2015
|
|
5IVS
 
 | |
4Y4N
 
 | | Thiazole synthase Thi4 from Methanococcus igneus | | Descriptor: | 2-[(E)-[(4R)-5-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-4-oxidanyl-3-oxidanylidene-pentan-2-ylidene]amino]ethanoic acid, FE (II) ION, Putative ribose 1,5-bisphosphate isomerase | | Authors: | Zhang, X, Ealick, S.E. | | Deposit date: | 2015-02-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Iron-Mediated Sulfur Transfer in Archael and Yeast Thiazole Synthases.
Biochemistry, 55, 2016
|
|
6UJB
 
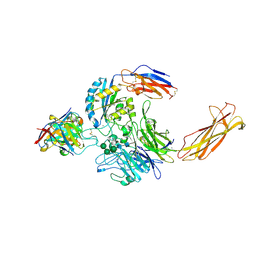 | | Integrin alpha-v beta-8 in complex with the Fabs C6D4 and 11D12v2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C6D4 heavy chain Fab, ... | | Authors: | Campbell, M.G, Cormier, A, Cheng, Y, Nishimura, S.L. | | Deposit date: | 2019-10-02 | | Release date: | 2020-02-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Cryo-EM Reveals Integrin-Mediated TGF-beta Activation without Release from Latent TGF-beta.
Cell, 180, 2020
|
|
5G4R
 
 | |
6UOX
 
 | | Structure of itraconazole-bound NPC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(3-bromo-4-{4-[4-({(2R,4S)-2-(2,4-dichlorophenyl)-2-[(1H-1,2,4-triazol-1-yl)methyl]-1,3-dioxolan-4-yl}methoxy)phenyl]piperazin-1-yl}phenyl)-2-[(2S)-butan-2-yl]-2,4-dihydro-3H-1,2,4-triazol-3-one, ... | | Authors: | Long, T, Li, X. | | Deposit date: | 2019-10-15 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Structural basis for itraconazole-mediated NPC1 inhibition.
Nat Commun, 11, 2020
|
|
5GWZ
 
 | | The structure of Porcine epidemic diarrhea virus main protease in complex with an inhibitor | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, PEDV main protease | | Authors: | Wang, F, Chen, C, Yang, K, Liu, X, Liu, H, Xu, Y, Chen, X, Liu, X, Cai, Y, Yang, H. | | Deposit date: | 2016-09-14 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Michael Acceptor-Based Peptidomimetic Inhibitor of Main Protease from Porcine Epidemic Diarrhea Virus
J. Med. Chem., 60, 2017
|
|
8DS8
 
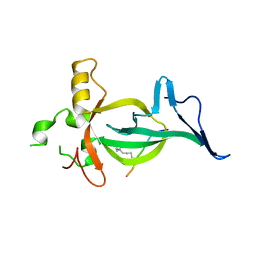 | |
7XW7
 
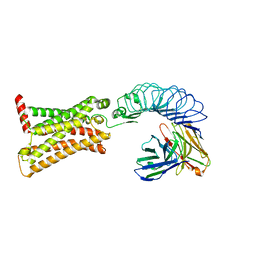 | | TSHR-K1-70 complex | | Descriptor: | K1-70 scFv, Thyrotropin receptor | | Authors: | Duan, J, Xu, P, Luan, X, Ji, Y, Yuan, Q, He, X, Ye, J, Cheng, X, Jiang, H, Zhang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-05-26 | | Release date: | 2022-08-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Hormone- and antibody-mediated activation of the thyrotropin receptor.
Nature, 609, 2022
|
|
5JR2
 
 | | Crystal structure of the EphA4 LBD in complex with APYd3 peptide inhibitor | | Descriptor: | APYd3 peptide, Ephrin type-A receptor 4, GLYCEROL, ... | | Authors: | Lechtenberg, B.C, Olson, E.J, Pasquale, E.B, Dawson, P.E, Riedl, S.J. | | Deposit date: | 2016-05-05 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Modifications of a Nanomolar Cyclic Peptide Antagonist for the EphA4 Receptor To Achieve High Plasma Stability.
Acs Med.Chem.Lett., 7, 2016
|
|
6FTZ
 
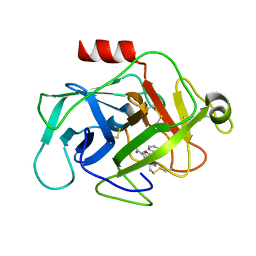 | | COMPLEMENT FACTOR D COMPLEXED WITH COMPOUND 6 | | Descriptor: | Complement factor D, ~{N}4-[3-(aminomethyl)phenyl]-1~{H}-indole-2,4-dicarboxamide | | Authors: | Ostermann, N. | | Deposit date: | 2018-02-26 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
6FUH
 
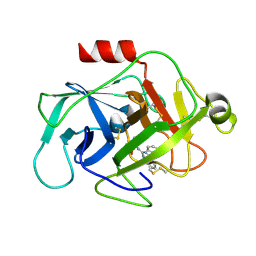 | | Complement factor D in complex with the inhibitor (4-((3-(aminomethyl)phenyl)amino)quinazolin-2-yl)-L-valine | | Descriptor: | (2~{S})-2-[[4-[[3-(aminomethyl)phenyl]amino]quinazolin-2-yl]amino]-3-methyl-butanoic acid, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
6FQO
 
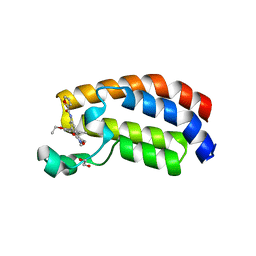 | | Crystal structure of CREBBP bromodomain complexd with DT29 | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein, ~{N}-[3-(2,5-dimethyl-3-oxidanylidene-1,2-oxazol-4-yl)-5-(5-ethanoyl-2-ethoxy-phenyl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
6FQU
 
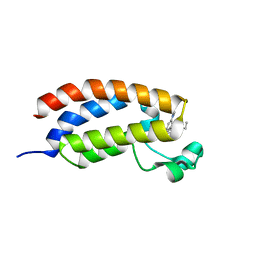 | | Crystal structure of CREBBP bromodomain complexd with DR09 | | Descriptor: | 1-[3-[3-[3,3-bis(fluoranyl)piperidin-1-yl]phenyl]-4-ethoxy-phenyl]ethanone, CREB-binding protein | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
6FUI
 
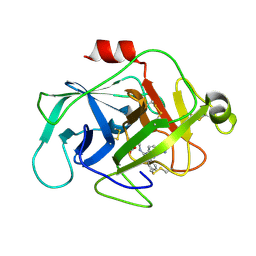 | | Complement factor D in complex with the inhibitor 3-((3-((3-(aminomethyl)phenyl)amino)-1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino)phenol | | Descriptor: | (1~{R},2~{S})-2-[[4-[[3-(aminomethyl)phenyl]amino]quinazolin-2-yl]amino]cyclohexane-1-carboxylic acid, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
5H8E
 
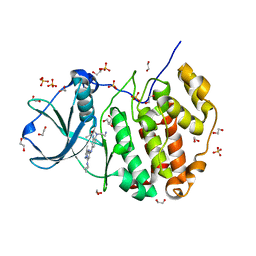 | | Crystal structure of CK2 with compound 7h | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent and Selective CK2 Kinase Inhibitors with Effects on Wnt Pathway Signaling in Vivo.
Acs Med.Chem.Lett., 7, 2016
|
|
5DYN
 
 | | B. fragilis cysteine protease | | Descriptor: | CHLORIDE ION, Putative peptidase, SODIUM ION | | Authors: | Choi, V.M, Herrou, J, Hecht, A.L, Turner, J.R, Crosson, S, Bubeck Wardenburg, J. | | Deposit date: | 2015-09-24 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Activation of Bacteroides fragilis toxin by a novel bacterial protease contributes to anaerobic sepsis in mice.
Nat. Med., 22, 2016
|
|
7PBL
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s1 [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBM
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s2 [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBO
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s4 [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
7PBT
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s1 [t1 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Wald, J, Fahrenkamp, D, Goessweiner-Mohr, N, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
6Q5A
 
 | |
7PBP
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s5 [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
