3OEI
 
 | | Crystal structure of Mycobacterium tuberculosis RelJK (Rv3357-Rv3358-RelBE3) | | Descriptor: | CITRATE ANION, RelJ (Antitoxin Rv3357), RelK (Toxin Rv3358) | | Authors: | Miallau, L, Cascio, D, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-08-12 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.145 Å) | | Cite: | Comparative proteomics identifies the cell-associated lethality of M. tuberculosis RelBE-like toxin-antitoxin complexes.
Structure, 21, 2013
|
|
3OF7
 
 | | The Crystal Structure of Prp20p from Saccharomyces cerevisiae and Its Binding Properties to Gsp1p and Histones | | Descriptor: | Regulator of chromosome condensation | | Authors: | Wu, F, Liu, Y, Zhu, Z, Huang, H, Ding, B, Wu, J, Shi, Y. | | Deposit date: | 2010-08-14 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9A crystal structure of Prp20p from Saccharomyces cerevisiae and its binding properties to Gsp1p and histones.
J.Struct.Biol., 174, 2011
|
|
3OFF
 
 | |
3OG4
 
 | | The crystal structure of human interferon lambda 1 complexed with its high affinity receptor in space group P21212 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin 28 receptor, alpha (Interferon, ... | | Authors: | Miknis, Z.J, Magracheva, E, Lei, W, Zdanov, A, Kotenko, S.V, Wlodawer, A. | | Deposit date: | 2010-08-16 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of the complex of human interferon-lambda1 with its high affinity receptor interferon-lambdaR1.
J.Mol.Biol., 404, 2010
|
|
3OF4
 
 | |
3O79
 
 | | Crystal Structure of Wild-type Rabbit PrP 126-230 | | Descriptor: | CHLORIDE ION, GLYCEROL, Rabbit PrP, ... | | Authors: | Sweeting, B, Chakrabartty, A, Pai, E.F. | | Deposit date: | 2010-07-30 | | Release date: | 2010-11-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Prion disease susceptibility is affected by beta-structure folding propensity and local side-chain interactions in PrP.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3O7T
 
 | | Crystal Structure of Cyclophilin A from Moniliophthora perniciosa | | Descriptor: | Cyclophilin A | | Authors: | Monzani, P.S, Pereira, H.M, Gramacho, K.P, Meirelles, F.V, Oliva, G, Cascardo, J.C.M. | | Deposit date: | 2010-07-31 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of apo-cyclophilin and bounded cyclosporine A from Moniliophthora perniciosa
To be Published
|
|
3OB2
 
 | |
3OBF
 
 | | Crystal structure of putative transcriptional regulator, IclR family; targeted domain 129...302 | | Descriptor: | Putative transcriptional regulator, IclR family | | Authors: | Chang, C, Tesar, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-06 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of putative transcriptional regulator, IclR family; targeted domain 129...302
To be Published
|
|
3OHA
 
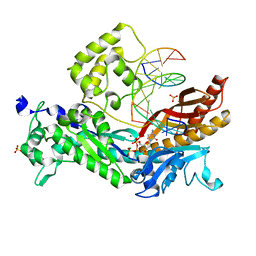 | | Yeast DNA polymerase eta inserting dCTP opposite an 8oxoG lesion | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*GP*TP*CP*CP*TP*CP*CP*CP*CP*TP*(DOC))-3', 5'-D(P*TP*(8OG)P*GP*AP*GP*GP*GP*GP*AP*GP*GP*AP*C)-3', ... | | Authors: | Silverstein, T.D, Jain, R, Aggarwal, A.K. | | Deposit date: | 2010-08-17 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for error-free replication of oxidatively damaged DNA by yeast DNA polymerase eta.
Structure, 18, 2010
|
|
3O66
 
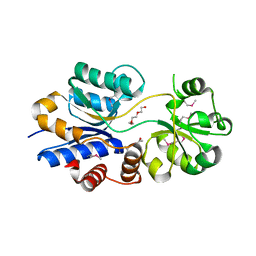 | | Crystal structure of glycine betaine/carnitine/choline ABC transporter | | Descriptor: | ACETATE ION, Glycine betaine/carnitine/choline ABC transporter, TRIETHYLENE GLYCOL | | Authors: | Chang, C, Bigelow, L, Carroll, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of glycine betaine/carnitine/choline ABC transporter
To be Published
|
|
3O6R
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with pyrogallol | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, BENZENE-1,2,3-TRIOL, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomitseva, M, Golovleva, L. | | Deposit date: | 2010-07-29 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
3O88
 
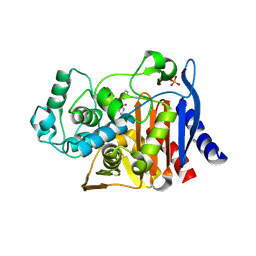 | | Crystal structure of AmpC beta-lactamase in complex with a sulfonamide boronic acid inhibitor | | Descriptor: | 3-[(2R)-2-[(benzylsulfonyl)amino]-2-(dihydroxyboranyl)ethyl]benzoic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Romagnoli, C, Karpiak, J, Shoichet, B.K. | | Deposit date: | 2010-08-02 | | Release date: | 2010-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Design, Synthesis, Crystal Structures, and Antimicrobial Activity of Sulfonamide Boronic Acids as beta-Lactamase Inhibitors
J.Med.Chem., 53, 2010
|
|
3O8S
 
 | |
3O8V
 
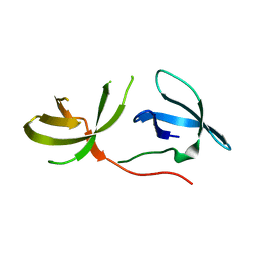 | | Crystal Structure of the Tudor Domains from FXR1 | | Descriptor: | Fragile X mental retardation syndrome-related protein 1 | | Authors: | Lam, R, Guo, Y.H, Adams-Cioaba, M, Bian, C.B, Mackenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-03 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of the Tandem Tudor Domains of Fragile X Mental Retardation Related Proteins FXR1 and FXR2.
Plos One, 5, 2010
|
|
3O9I
 
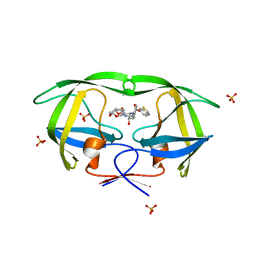 | | Crystal Structure of wild-type HIV-1 Protease in complex with af61 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-3-[(1,3-benzothiazol-6-ylsulfonyl)(2-ethylbutyl)amino]-1-benzyl-2-hydroxypropyl}carbamate, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
3OAC
 
 | | Mint deletion mutant of heterotetrameric geranyl pyrophosphate synthase in complex with ligands | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Geranyl diphosphate synthase large subunit, ... | | Authors: | Hsieh, F.-L, Chang, T.-H, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2010-08-05 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Enhanced specificity of mint geranyl pyrophosphate synthase by modifying the R-loop interactions
J.Mol.Biol., 404, 2010
|
|
3O9D
 
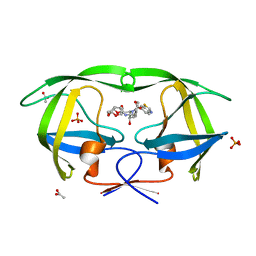 | | Crystal Structure of wild-type HIV-1 Protease in complex with kd19 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{(1,3-benzothiazol-6-ylsulfonyl)[(2S)-2-methylbutyl]amino}-1-benzyl-2-hydroxypropyl]carbamate, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2010-08-04 | | Release date: | 2011-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance.
Chem.Biol., 20, 2013
|
|
3OB0
 
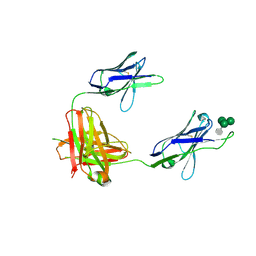 | | A non-self sugar mimic of the HIV glycan shield shows enhanced antigenicity | | Descriptor: | 7-deoxy-L-glycero-alpha-D-manno-heptopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose, Fab 2G12, heavy chain, ... | | Authors: | Doores, K.J, Fulton, Z, Hong, V, Patel, M.K, Scanlan, C.N, Wormald, M.R, Finn, M.G, Burton, D.R, Wilson, I.A, Davis, B.G. | | Deposit date: | 2010-08-06 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OBT
 
 | |
3ODA
 
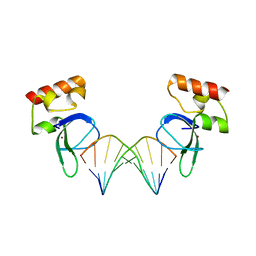 | | Human PARP-1 zinc finger 1 (Zn1) bound to DNA | | Descriptor: | 5'-D(*GP*CP*CP*TP*GP*CP*AP*GP*GP*C)-3', Poly [ADP-ribose] polymerase 1, ZINC ION | | Authors: | Pascal, J.M, Langelier, M.-F. | | Deposit date: | 2010-08-11 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal Structures of Poly(ADP-ribose) Polymerase-1 (PARP-1) Zinc Fingers Bound to DNA: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO DNA-DEPENDENT PARP-1 ACTIVITY.
J.Biol.Chem., 286, 2011
|
|
3OCY
 
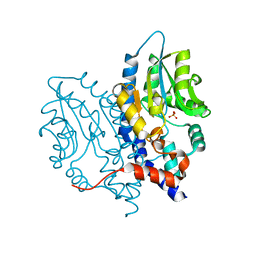 | | Structure of Recombinant Haemophilus Influenzae e(P4) Acid Phosphatase Complexed with inorganic phosphate | | Descriptor: | Lipoprotein E, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | Deposit date: | 2010-08-10 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Recognition of nucleoside monophosphate substrates by Haemophilus influenzae class C acid phosphatase.
J.Mol.Biol., 404, 2010
|
|
3OD9
 
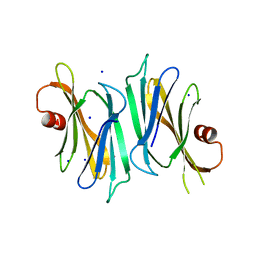 | | Crystal structure of PliI-Ah, periplasmic lysozyme inhibitor of I-type lysozyme from Aeromonas hydrophyla | | Descriptor: | POTASSIUM ION, Putative exported protein, SODIUM ION | | Authors: | Leysen, S, Van Herreweghe, J.M, Callewaert, L, Heirbaut, M, Buntinx, P, Michiels, C.W, Strelkov, S.V. | | Deposit date: | 2010-08-11 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.411 Å) | | Cite: | Molecular Basis of Bacterial Defense against Host Lysozymes: X-ray Structures of Periplasmic Lysozyme Inhibitors PliI and PliC.
J.Mol.Biol., 405, 2011
|
|
3ODP
 
 | |
3OE1
 
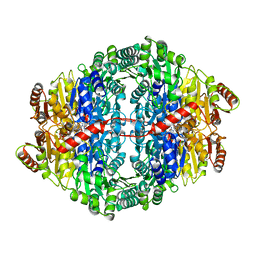 | | Pyruvate decarboxylase variant Glu473Asp from Z. mobilis in complex with reaction intermediate 2-lactyl-ThDP | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Meyer, D, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Double duty for a conserved glutamate in pyruvate decarboxylase: evidence of the participation in stereoelectronically controlled decarboxylation and in protonation of the nascent carbanion/enamine intermediate .
Biochemistry, 49, 2010
|
|
