3C6G
 
 | | Crystal structure of CYP2R1 in complex with vitamin D3 | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), Cytochrome P450 2R1, ... | | Authors: | Strushkevich, N.V, Min, J, Loppnau, P, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Plotnikov, A.N, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural analysis of CYP2R1 in complex with vitamin D3.
J.Mol.Biol., 380, 2008
|
|
7LSO
 
 | | L-Phenylseptin | | Descriptor: | L-Phenylseptin peptide | | Authors: | Munhoz, V.H, Verly, R.M. | | Deposit date: | 2021-02-18 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of L-Phenylseptin in DPC-d38.
To Be Published
|
|
7LSP
 
 | |
1PS9
 
 | | The Crystal Structure and Reaction Mechanism of E. coli 2,4-Dienoyl CoA Reductase | | Descriptor: | 2,4-dienoyl-CoA reductase, 5-MERCAPTOETHANOL-2-DECENOYL-COENZYME A, CHLORIDE ION, ... | | Authors: | Hubbard, P.A, Liang, X, Schulz, H, Kim, J.J. | | Deposit date: | 2003-06-20 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure and reaction mechanism of Escherichia coli 2,4-dienoyl-CoA reductase
J.Biol.Chem., 278, 2003
|
|
3BSX
 
 | | Crystal Structure of Human Pumilio 1 in complex with Puf5 RNA | | Descriptor: | 5'-R(*UP*UP*GP*UP*AP*AP*UP*AP*UP*UP*A)-3', Pumilio homolog 1 | | Authors: | Gupta, Y.K, Nair, D.T, Wharton, R.P, Aggarwal, A.K. | | Deposit date: | 2007-12-26 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity.
Structure, 16, 2008
|
|
7LSY
 
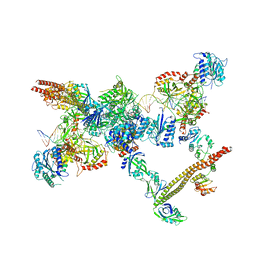 | | NHEJ Short-range synaptic complex | | Descriptor: | DNA (26-MER), DNA (5'-D(P*CP*AP*AP*TP*GP*AP*AP*AP*CP*GP*GP*AP*AP*CP*AP*GP*TP*CP*AP*G)-3'), DNA (5'-D(P*GP*TP*TP*CP*TP*TP*AP*GP*TP*AP*TP*AP*TP*A)-3'), ... | | Authors: | He, Y, Chen, S. | | Deposit date: | 2021-02-18 | | Release date: | 2021-04-14 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structural basis of long-range to short-range synaptic transition in NHEJ.
Nature, 593, 2021
|
|
4LY6
 
 | | Nucleotide-induced asymmetry within ATPase activator ring drives s54-RNAP interaction and ATP hydrolysis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transcriptional regulator (NtrC family), ... | | Authors: | Sysoeva, T.A, Chowdhury, S, Guo, L, Nixon, B.T. | | Deposit date: | 2013-07-30 | | Release date: | 2013-12-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Nucleotide-induced asymmetry within ATPase activator ring drives sigma 54-RNAP interaction and ATP hydrolysis.
Genes Dev., 27, 2013
|
|
5H77
 
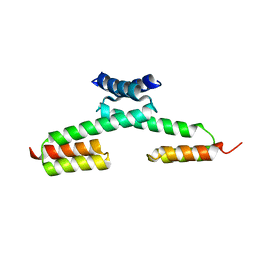 | | Crystal structure of the PKA-protein A fusion protein | | Descriptor: | cAMP-dependent protein kinase type II-alpha regulatory subunit,Immunoglobulin G-binding protein A | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2016-11-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.197 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
7M1M
 
 | | Crystal structure of Pseudomonas aeruginosa ClpP1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | Authors: | Mawla, G.D, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa.
Mol.Microbiol., 115, 2021
|
|
3C78
 
 | |
2VN0
 
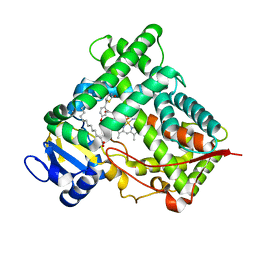 | | CYP2C8DH COMPLEXED WITH TROGLITAZONE | | Descriptor: | (5R)-5-(4-{[(2R)-6-HYDROXY-2,5,7,8-TETRAMETHYL-3,4-DIHYDRO-2H-CHROMEN-2-YL]METHOXY}BENZYL)-1,3-THIAZOLIDINE-2,4-DIONE, CYTOCHROME P450 2C8, PALMITIC ACID, ... | | Authors: | Schoch, G.A, Yano, J.K, Sansen, S, Stout, C.D, Johnson, E.F. | | Deposit date: | 2008-01-30 | | Release date: | 2008-04-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Determinants of cytochrome P450 2C8 substrate binding: structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid.
J. Biol. Chem., 283, 2008
|
|
3BYW
 
 | | Crystal structure of an extracellular domain of arabinofuranosyltransferase from Corynebacterium diphtheriae | | Descriptor: | ACETATE ION, Putative arabinofuranosyltransferase, ZINC ION | | Authors: | Tan, K, Hatzos, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of an extracellular domain of arabinofuranosyltransferase from Corynebacterium diphtheriae.
To be Published
|
|
7KF4
 
 | | Crystal structure from SARS-CoV-2 NendoU NSP15 | | Descriptor: | CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Nakamura, A.M, Pereira, H.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliveira, K.I.Z, Oliva, G. | | Deposit date: | 2020-10-13 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
4MEX
 
 | | Crystal structure of Escherichia coli RNA polymerase in complex with salinamide A | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Feng, Y, Zhang, Y, Arnold, E, Ebright, R.H. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-21 | | Last modified: | 2014-06-25 | | Method: | X-RAY DIFFRACTION (3.902 Å) | | Cite: | Transcription inhibition by the depsipeptide antibiotic salinamide A.
Elife, 3, 2014
|
|
1Q5Q
 
 | | The Rhodococcus 20S proteasome | | Descriptor: | proteasome alpha-type subunit 1, proteasome beta-type subunit 1 | | Authors: | Kwon, Y.D, Nagy, I, Adams, P.D, Baumeister, W, Jap, B.K. | | Deposit date: | 2003-08-08 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the Rhodococcus proteasome with and without its pro-peptides: implications for the role of the pro-peptide in proteasome assembly.
J.Mol.Biol., 335, 2004
|
|
7KQY
 
 | | Crystal Structure and Characterization of Human Heavy-Chain only Antibodies reveals a novel, stable dimeric structure similar to Monoclonal Antibodies | | Descriptor: | Heavy-Chain only Human Antibodies | | Authors: | Bahmanjah, S, Mieczkowski, C, Yu, Y, Baker, J, Raghunathan, G, Tomazela, D, Hsieh, M, Mccoy, M, Strickland, C, Fayadat-Dilman, L. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Crystal Structure and Characterization of Human Heavy-Chain Only Antibodies Reveals a Novel, Stable Dimeric Structure Similar to Monoclonal Antibodies.
Antibodies, 9, 2020
|
|
7KSO
 
 | | Cryo-EM structure of PRC2:EZH1-AEBP2-JARID2 | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, ... | | Authors: | Grau, D.J, Armache, K.J. | | Deposit date: | 2020-11-23 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
3C60
 
 | |
7KSR
 
 | | PRC2:EZH1_A from a dimeric PRC2 bound to a nucleosome | | Descriptor: | Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH1, Polycomb protein EED, ... | | Authors: | Grau, D.J, Armache, K.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of monomeric and dimeric PRC2:EZH1 reveal flexible modules involved in chromatin compaction.
Nat Commun, 12, 2021
|
|
7KHN
 
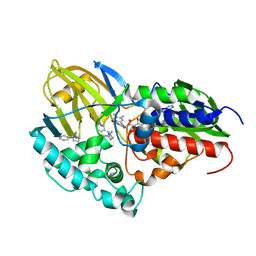 | | NicA2 variant N462Y/W427Y in complex with (S)-nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, Amine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Tararina, M.A, Allen, K.N. | | Deposit date: | 2020-10-21 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fast Kinetics Reveals Rate-Limiting Oxidation and the Role of the Aromatic Cage in the Mechanism of the Nicotine-Degrading Enzyme NicA2.
Biochemistry, 60, 2021
|
|
7KZO
 
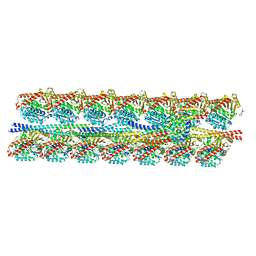 | | Outer dynein arm docking complex bound to doublet microtubules from C. reinhardtii | | Descriptor: | Dynein gamma chain, flagellar outer arm, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Walton, T, Wu, H, Brown, A.B. | | Deposit date: | 2020-12-10 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a microtubule-bound axonemal dynein.
Nat Commun, 12, 2021
|
|
4MDH
 
 | |
5GKZ
 
 | | Structure of RyR1 in a closed state (C3 conformer) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ZINC ION | | Authors: | Bai, X.C, Yan, Z, Wu, J.P, Yan, N. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The Central domain of RyR1 is the transducer for long-range allosteric gating of channel opening
Cell Res., 26, 2016
|
|
3CAU
 
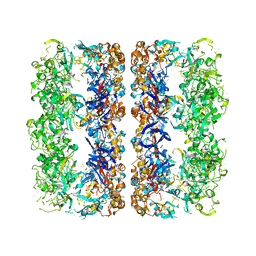 | | D7 symmetrized structure of unliganded GroEL at 4.2 Angstrom resolution by cryoEM | | Descriptor: | 60 kDa chaperonin | | Authors: | Ludtke, S.J, Baker, M.L, Chen, D.H, Song, J.L, Chuang, D, Chiu, W. | | Deposit date: | 2008-02-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | De Novo Backbone Trace of GroEL from Single Particle Electron Cryomicroscopy.
Structure, 16, 2008
|
|
3FWC
 
 | | Sac3:Sus1:Cdc31 complex | | Descriptor: | Cell division control protein 31, Nuclear mRNA export protein SAC3, Protein SUS1, ... | | Authors: | Stewart, M, Jani, D. | | Deposit date: | 2009-01-17 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export.
Mol.Cell, 33, 2009
|
|
