3F3K
 
 | | The structure of uncharacterized protein YKR043C from Saccharomyces cerevisiae. | | Descriptor: | GLYCEROL, Uncharacterized protein YKR043C | | Authors: | Cuff, M, Xu, X, Cui, H, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae.
J.Biol.Chem., 285, 2010
|
|
3F41
 
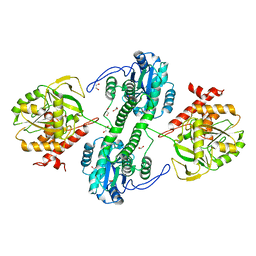 | | Structure of the tandemly repeated protein tyrosine phosphatase like phytase from Mitsuokella multacida | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Phytase | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2008-10-31 | | Release date: | 2009-06-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of a multifunctional, tandemly repeated inositol polyphosphatase.
J.Mol.Biol., 392, 2009
|
|
3F46
 
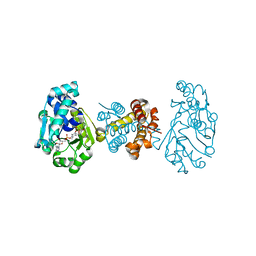 | | The Crystal Structure of C176A Mutated [Fe]-Hydrogenase (Hmd) Holoenzyme from Methanocaldococcus jannaschii | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 5'-O-[(S)-hydroxy{[2-hydroxy-3,5-dimethyl-6-(2-oxoethyl)pyridin-4-yl]oxy}phosphoryl]guanosine, 5,10-methenyltetrahydromethanopterin hydrogenase, ... | | Authors: | Hiromoto, T, Vogt, S, Warkentin, E, Thauer, R.K, Shima, S, Ermler, U. | | Deposit date: | 2008-10-31 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of C176A mutated [Fe]-hydrogenase suggests an acyl-iron ligation in the active site iron complex.
Febs Lett., 583, 2009
|
|
3F4W
 
 | |
3NYL
 
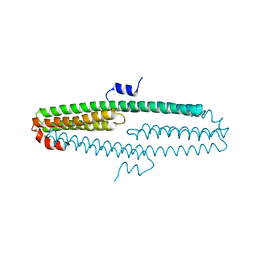 | | The X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain | | Descriptor: | Amyloid beta (A4) protein (Peptidase nexin-II, Alzheimer disease), isoform CRA_b | | Authors: | Ha, Y, Hu, J, Lee, S, Liu, X, Wang, Y. | | Deposit date: | 2010-07-15 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain.
Mol.Cell, 15, 2004
|
|
3F5T
 
 | | X-ray Structure of H5N1 NS1 | | Descriptor: | Nonstructural protein 1 | | Authors: | Bornholdt, Z.A, Prasad, B.V.V. | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus
Nature, 456, 2008
|
|
3O6J
 
 | | Crystal Structure of 4-Chlorocatechol Dioxygenase from Rhodococcus opacus 1CP in complex with hydroxyquinol | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, CHLORIDE ION, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Briganti, F, Kolomitseva, M, Golovleva, L. | | Deposit date: | 2010-07-29 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray structures of 4-chlorocatechol 1,2-dioxygenase adducts with substituted catechols: new perspectives in the molecular basis of intradiol ring cleaving dioxygenases specificity.
J. Struct. Biol., 181, 2013
|
|
3FJV
 
 | |
3OAB
 
 | | Mint deletion mutant of heterotetrameric geranyl pyrophosphate synthase in complex with ligands | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | Authors: | Hsieh, F.-L, Chang, T.-H, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2010-08-05 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enhanced specificity of mint geranyl pyrophosphate synthase by modifying the R-loop interactions
J.Mol.Biol., 404, 2010
|
|
3OAZ
 
 | | A non-self sugar mimic of the HIV glycan shield shows enhanced antigenicity | | Descriptor: | CHLORIDE ION, Fab 2G12, heavy chain, ... | | Authors: | Doores, K.J, Fulton, Z, Hong, V, Patel, M.K, Scanlan, C.N, Wormald, M.R, Finn, M.G, Burton, D.R, Wilson, I.A, Davis, B.G. | | Deposit date: | 2010-08-06 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A nonself sugar mimic of the HIV glycan shield shows enhanced antigenicity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NO6
 
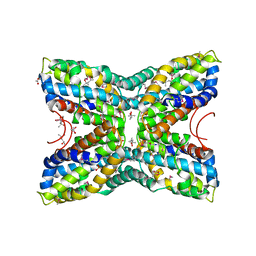 | |
3OBV
 
 | | Autoinhibited Formin mDia1 Structure | | Descriptor: | Protein diaphanous homolog 1, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Tomchick, D.R, Rosen, M.K, Otomo, T. | | Deposit date: | 2010-08-09 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the Formin mDia1 in autoinhibited conformation.
Plos One, 5, 2010
|
|
3NQI
 
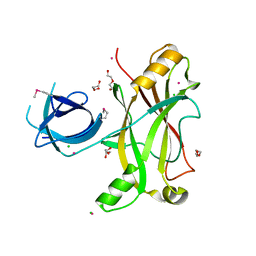 | |
3EYH
 
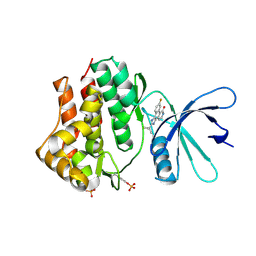 | | Crystal structures of JAK1 and JAK2 inhibitor complexes | | Descriptor: | 2-TERT-BUTYL-9-FLUORO-3,6-DIHYDRO-7H-BENZ[H]-IMIDAZ[4,5-F]ISOQUINOLINE-7-ONE, Tyrosine-protein kinase | | Authors: | Williams, N.K, Bamert, R.S, Patell, O, Wang, C, Walden, P.M, Fantino, E. | | Deposit date: | 2008-10-20 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dissecting specificity in the Janus kinases: the structures of JAK-specific inhibitors complexed to the JAK1 and JAK2 protein tyrosine kinase domains.
J.Mol.Biol., 387, 2009
|
|
3NRA
 
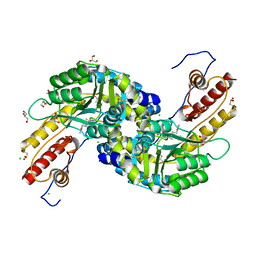 | |
3EZ0
 
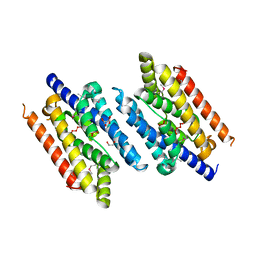 | |
3NTS
 
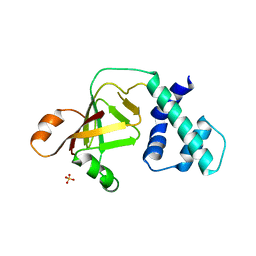 | |
3F8X
 
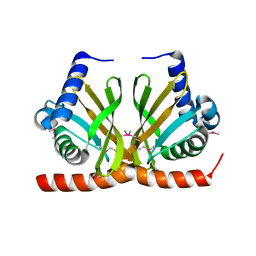 | |
3F9F
 
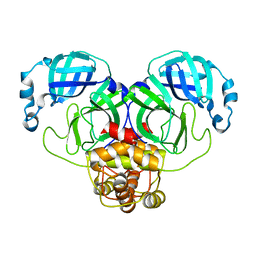 | |
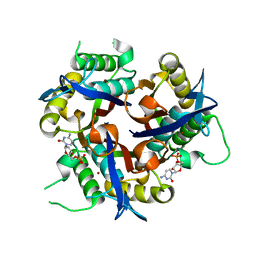
3F0G
 
 | |
3FA1
 
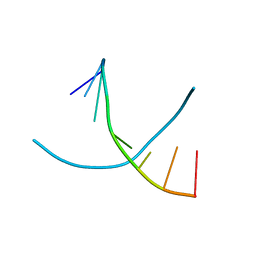 | |
3NV9
 
 | | Crystal structure of Entamoeba histolytica Malic Enzyme | | Descriptor: | ACETATE ION, GLYCEROL, Malic enzyme, ... | | Authors: | Dutta, D, Chakrabarty, A, Ghosh, S.K, Das, A.K. | | Deposit date: | 2010-07-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional characterization of Entamoeba histolytica Malic enzyme
To be Published
|
|
3FBI
 
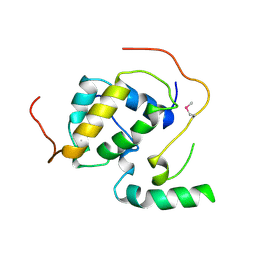 | | Structure of the Mediator submodule Med7N/31 | | Descriptor: | Mediator of RNA polymerase II transcription subunit 31, Mediator of RNA polymerase II transcription subunit 7 | | Authors: | Koschubs, T, Seizl, M, Lariviere, L, Kurth, F, Baumli, S, Martin, D.E, Cramer, P. | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification, structure, and functional requirement of the Mediator submodule Med7N/31
Embo J., 28, 2009
|
|
3NYO
 
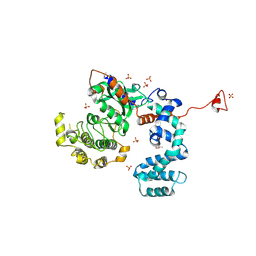 | | Crystal Structure of G Protein-Coupled Receptor Kinase 6 in Complex with AMP | | Descriptor: | (R,R)-2,3-BUTANEDIOL, ADENOSINE MONOPHOSPHATE, G protein-coupled receptor kinase 6, ... | | Authors: | Tesmer, J.J.G, Singh, P. | | Deposit date: | 2010-07-15 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Molecular basis for activation of G protein-coupled receptor kinases.
Embo J., 29, 2010
|
|
3NRD
 
 | |
